A dose-dependent perturbation in cardiac energy metabolism is linked to radiation-induced ischemic heart disease in Mayak nuclear workers - PubMed (original) (raw)
A dose-dependent perturbation in cardiac energy metabolism is linked to radiation-induced ischemic heart disease in Mayak nuclear workers
Omid Azimzadeh et al. Oncotarget. 2017.
Abstract
Epidemiological studies show a significant increase in ischemic heart disease (IHD) incidence associated with total external gamma-ray dose among Mayak plutonium enrichment plant workers. Our previous studies using mouse models suggest that persistent alteration of heart metabolism due to the inhibition of peroxisome proliferator-activated receptor (PPAR) alpha accompanies cardiac damage after high doses of ionising radiation. The aim of the present study was to elucidate the mechanism of radiation-induced IHD in humans. The cardiac proteome response to irradiation was analysed in Mayak workers who were exposed only to external doses of gamma rays. All participants were diagnosed during their lifetime with IHD that also was the cause of death. Label-free quantitative proteomics analysis was performed on tissue samples from the cardiac left ventricles of individuals stratified into four radiation dose groups (0 Gy, < 100 mGy, 100-500 mGy, and > 500 mGy). The groups could be separated using principal component analysis based on all proteomics features. Proteome profiling showed a dose-dependent increase in the number of downregulated mitochondrial and structural proteins. Both proteomics and immunoblotting showed decreased expression of several oxidative stress responsive proteins in the irradiated hearts. The phosphorylation of transcription factor PPAR alpha was increased in a dose-dependent manner, which is indicative of a reduction in transcriptional activity with increased radiation dose. These data suggest that chronic external radiation enhances the risk for IHD by inhibiting PPAR alpha and altering the expression of mitochondrial, structural, and antioxidant components of the heart.
Keywords: PPAR alpha; heart disease; ionising radiation; mitochondrial dysfunction; proteomics.
Conflict of interest statement
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Figures
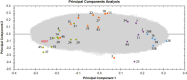
Figure 1. Principal component analysis (PCA) based on all proteomic features in the left ventricle of sample donors in different dose groups
The PCA used features with charges +2 to +7 resulting in PC1 and PC2 as follows: PC1 23.65% and PC2 8.36%. The control samples with the corresponding donor number are represented as blue spots, the samples exposed to < 100 mGy in purple, the samples exposed to 100–500 mGy in orange and the samples exposed to > 500 mGy in green. Samples number 26 and 38 were run as 2 technical replicates and are indicated as 26, 26B and 38, 38B. Detailed information of the sample donors and the exact doses are given in Supplementary Table S10. The analysis was performed using the Progenesis QI software (
).
Figure 2. Pathway and network analysis of significantly differentially expressed mitochondrial proteins
A dose-dependent alteration is observed in the pathways involved in the energy production. The pathway scores are displayed using a purple colour gradient, where darker purple corresponds to higher scores (increased statistical significance). The score is the negative log of the _p_-value derived from the Fisher′s Exact test. By default, the rows (pathways) with the highest total score across the set of observations are sorted to the top (A). Heat map for the expression values of differentially expressed OXPHOS proteins between dose groups is displayed using a green colour gradient for downregulated proteins, where dark green corresponds to large downregulation. The numbers shows how many proteins were deregulated in each subunit (B) (
). Protein-protein interaction analysis of the significantly differentially expressed proteins showing the networks of deregulated mitochondrial proteins in the dose groups < 100 mGy (**C**), 100–500 mGy (**D**) and > 500 mGy (E) (
).
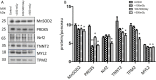
Figure 3. Immunoblot validation of the proteomics data
The heart protein lysates from each individual sample were pooled within the dose groups and tested using anti-Troponin T (TNNT2), anti-Tropomyosin 2 (TPM2), anti- Myosin light chain (MYL2), anti-Mn superoxide dismutase (SOD2), and anti-Peroxiredoxin 5 (PRDX5) (A).The columns represent the average ratios of relative protein expression in control and irradiated samples. The amount of the total protein was measured by Ponceau S staining for accurate comparison between the groups. The error bars represent standard error of the mean (+ SEM) (B) (_t_-test; *p < 0.05, **p < 0.01; n = 3).
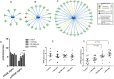
Figure 4. Analysis of the protein carbonyl levels and miR-21 and miR-146a in different dose groups
The total amount of carbonylated protein was measured in individual samples from each dose group. The samples in the control group were run in two technical replicates. Significantly increased level of carbonylated proteins was shown in the dose group > 500 mGy (A). Analysis of miR-21 and miR-146a from samples of all dose groups showed significant upregulation of both miRNAs in the dose group > 500 mGy (B) The error bars represent standard error of the mean (+ SEM) (_t_-test; * p < 0.05; **p < 0.01; *** p < 0.001).
Figure 5. Analysis of the activation status of PPAR alpha
IPA prediction of inactivation of PPAR alpha based on the deregulated proteins from proteomics analysis in the dose groups < 100 mGy (**A**), 100–500 mGy (**B**) and > 500 mGy (C). The upregulated proteins are marked in red and the down-regulated in green. The blue colour of the node (PPAR alpha) indicates inactivation. The list of proteins is available in Supplementary Tables S2–S4. Immunoblot analysis of total and phospho-PPAR alpha (Ser12) in pooled samples is shown (D). The columns represent the average ratios of relative protein expression in control and irradiated samples. Immunoblot analysis of total PPAR alpha (E) and phospho-PPAR alpha (F) in individual samples from each dose group is shown. The icons represent individual samples in different dose groups. The samples in the control group were run in two technical replicates. The amount of the total protein was confirmed by Ponceau S staining for accurate comparison between the groups (_t_-test; *p < 0.05).
Similar articles
- Chronic Occupational Exposure to Ionizing Radiation Induces Alterations in the Structure and Metabolism of the Heart: A Proteomic Analysis of Human Formalin-Fixed Paraffin-Embedded (FFPE) Cardiac Tissue.
Azimzadeh O, Azizova T, Merl-Pham J, Blutke A, Moseeva M, Zubkova O, Anastasov N, Feuchtinger A, Hauck SM, Atkinson MJ, Tapio S. Azimzadeh O, et al. Int J Mol Sci. 2020 Sep 17;21(18):6832. doi: 10.3390/ijms21186832. Int J Mol Sci. 2020. PMID: 32957660 Free PMC article. - Integrative multiomics study for validation of mechanisms in radiation-induced ischemic heart disease in Mayak workers.
Papiez A, Azimzadeh O, Azizova T, Moseeva M, Anastasov N, Smida J, Tapio S, Polanska J. Papiez A, et al. PLoS One. 2018 Dec 31;13(12):e0209626. doi: 10.1371/journal.pone.0209626. eCollection 2018. PLoS One. 2018. PMID: 30596717 Free PMC article. - Hyperacetylation of Cardiac Mitochondrial Proteins Is Associated with Metabolic Impairment and Sirtuin Downregulation after Chronic Total Body Irradiation of ApoE -/- Mice.
Barjaktarovic Z, Merl-Pham J, Braga-Tanaka I, Tanaka S, Hauck SM, Saran A, Mancuso M, Atkinson MJ, Tapio S, Azimzadeh O. Barjaktarovic Z, et al. Int J Mol Sci. 2019 Oct 22;20(20):5239. doi: 10.3390/ijms20205239. Int J Mol Sci. 2019. PMID: 31652604 Free PMC article. - Radiation in the workplace-a review of studies of the risks of occupational exposure to ionising radiation.
Wakeford R. Wakeford R. J Radiol Prot. 2009 Jun;29(2A):A61-79. doi: 10.1088/0952-4746/29/2A/S05. Epub 2009 May 19. J Radiol Prot. 2009. PMID: 19454806 Review. - Quantitative comparisons of cancer induction in humans by internally deposited radionuclides and external radiation.
Harrison JD, Muirhead CR. Harrison JD, et al. Int J Radiat Biol. 2003 Jan;79(1):1-13. Int J Radiat Biol. 2003. PMID: 12556326 Review.
Cited by
- Quantitative Proteomic Analysis Using Formalin-Fixed, Paraffin-Embedded (FFPE) Human Cardiac Tissue.
Azimzadeh O, Atkinson MJ, Tapio S. Azimzadeh O, et al. Methods Mol Biol. 2021;2261:525-533. doi: 10.1007/978-1-0716-1186-9_33. Methods Mol Biol. 2021. PMID: 33421012 - Chronic Occupational Exposure to Ionizing Radiation Induces Alterations in the Structure and Metabolism of the Heart: A Proteomic Analysis of Human Formalin-Fixed Paraffin-Embedded (FFPE) Cardiac Tissue.
Azimzadeh O, Azizova T, Merl-Pham J, Blutke A, Moseeva M, Zubkova O, Anastasov N, Feuchtinger A, Hauck SM, Atkinson MJ, Tapio S. Azimzadeh O, et al. Int J Mol Sci. 2020 Sep 17;21(18):6832. doi: 10.3390/ijms21186832. Int J Mol Sci. 2020. PMID: 32957660 Free PMC article. - Ionizing radiation-induced circulatory and metabolic diseases.
Tapio S, Little MP, Kaiser JC, Impens N, Hamada N, Georgakilas AG, Simar D, Salomaa S. Tapio S, et al. Environ Int. 2021 Jan;146:106235. doi: 10.1016/j.envint.2020.106235. Epub 2020 Nov 3. Environ Int. 2021. PMID: 33157375 Free PMC article. Review. - The Role of Mitochondrial Dysfunction in Radiation-Induced Heart Disease: From Bench to Bedside.
Livingston K, Schlaak RA, Puckett LL, Bergom C. Livingston K, et al. Front Cardiovasc Med. 2020 Feb 21;7:20. doi: 10.3389/fcvm.2020.00020. eCollection 2020. Front Cardiovasc Med. 2020. PMID: 32154269 Free PMC article. Review. - The Molecular Mechanisms in Senescent Cells Induced by Natural Aging and Ionizing Radiation.
Ibragimova M, Kussainova A, Aripova A, Bersimbaev R, Bulgakova O. Ibragimova M, et al. Cells. 2024 Mar 21;13(6):550. doi: 10.3390/cells13060550. Cells. 2024. PMID: 38534394 Free PMC article. Review.
References
- Azizova TV, Muirhead CR, Druzhinina MB, Grigoryeva ES, Vlasenko EV, Sumina MV, O’Hagan JA, Zhang W, Haylock RG, Hunter N. Cardiovascular diseases in the cohort of workers first employed at Mayak PA in 1948–1958. Radiat Res. 2010;174:155–168. - PubMed
- Yamada M, Wong FL, Fujiwara S, Akahoshi M, Suzuki G. Noncancer disease incidence in atomic bomb survivors, 1958–1998. Radiat Res. 2004;161:622–632. - PubMed
- Stanley WC, Hoppel CL. Mitochondrial dysfunction in heart failure: potential for therapeutic interventions? Cardiovasc Res. 2000;45:805–806. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical