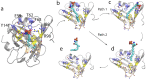Binding Characteristics of Sphingosine-1-Phosphate to ApoM hints to Assisted Release Mechanism via the ApoM Calyx-Opening - PubMed (original) (raw)
Binding Characteristics of Sphingosine-1-Phosphate to ApoM hints to Assisted Release Mechanism via the ApoM Calyx-Opening
Hansi Zhang et al. Sci Rep. 2016.
Abstract
Sphingosine-1-phosphate (S1P) is a lysophospholipid mediator carried by the HDL-associated apoM protein in blood, regulating many physiological processes by activating the G protein-coupled S1P receptor in mammals. Despite the solved crystal structure of the apoM-S1P complex, the mechanism of S1P release from apoM as a part of the S1P pathway is unknown. Here, the dynamics of the wild type apoM-S1P complex as well as of mutants were investigated by means of atomistic molecular dynamics simulations. The potential of mean force for S1P unbinding from apoM reflected a large binding strength of more than 60 kJ/mol. This high unbinding free energy for S1P underlines the observed specificity of the physiological effects of S1P as it suggests that the spontaneous release of S1P from apoM is unlikely. Instead, S1P release and thus the control of this bioactive lipid probably requires the tight interaction with other molecules, e.g. with the S1P receptor. Mutations of specific S1P anchoring residues of apoM decreased the energetic barrier by up to 20 kJ/mol. Moreover, the ligand-free apoM protein is shown to adopt a more open upper hydrophilic binding pocket and to result in complete closure of the lower hydrophobic cavity, suggesting a mechanism for adjusting the gate for ligand access.
Figures
Figure 1
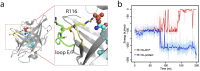
(a) The conformational switch of Arg116 after 80 ns of simulation time. (b) The interaction energy computed as the sum of short range Lennard-Jones and electrostatic interactions between Arg116 and S1P (red) and between Arg116 and the protein (blue).
Figure 2. Pairwise interaction forces between apoM and S1P.
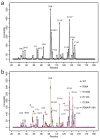
(a) Interaction forces for the apoM-S1P complex. The pairwise forces between S1P and each residue of apoM were calculated using the force distribution analysis (FDA, see Methods). The forces were averaged over simulations of the apoM WT both in solution and anchored to POPC. The residues with largest forces (>70 pN) are labeled. The four residues with the highest values (marked by asterisk) are in accordance with the S1P binding residues reported in the crystal structure. (b) Interaction forces during enforced S1P unbinding. Each line was averaged over three pulling simulations for each complex. Residues with forces >40 pN are marked. Residues with forces significantly exceeding those from wt equilibrium MD simulation are labeled in red and marked with an asterisk.
Figure 3. The S1P binding pocket of apoM.
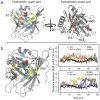
(a) A snapshot of wt apoM-S1P after 200 ns of equilibration. Left panel: The residues highlighted in yellow mainly contribute to the upper hydrophilic part of the binding pocket and stabilize the charged moiety of S1P. Right panel: The residues colored orange form the hydrophobic binding groove of apoM. (b) Three main conformational changes were observed for the equilibrium MD simulations of the studied mutants (colored green):① Arg116 switched to stabilize S1P in the R98A mutant, and occasionally also in the W100G and E136A mutants; ② The flexibility of Arg98 increased in the W100G mutant due to the loss of the cation-π interaction; ③ The distance between C_α_ atoms of residues 98 and 136 indicating calyx structure loosening increased for all mutants as compared to the WT protein. (c) The distances between C_α_ atoms of residues 98 and 136 as a function of simulation time for the WT (black), the WT anchored to a POPC bilayer (brown), the R98A (red), W100G (green), R116A (blue), E136A (orange), and R98A/R116A mutants (magenta).
Figure 4. Stability and fluctuations of apoM and S1P.
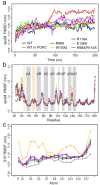
RMSD (a) and RMSF (b) values of apoM were analyzed for the C_α_-atoms of the seven studied complexes (WT, black; WT attached to POPC, brown; R98A, red; W100G, green; R116A, blue; E136A, orange; R98A/R116A, magenta). The values were averaged over 40 windows of 5 ns length each. Error bars show the standard error. (c) RMSF values of the phosphorus atom and carbon atoms of S1P. The values were averaged over 20 windows of 10 ns length each. Error bars show the standard error.
Figure 5
(a) Sequence of human apoM along with the secondary structure elements gained from the crystal structure. (b) S1P structure. Oxygen atoms, phosphorus atom, and nitrogen atom are shown in red, yellow and blue, respectively. (c) ApoM structure from residue 27 to residue 187, containing a _β_-barrel with eight anti-parallel _β_-strands displayed in cartoon representation. (d) The S1P-bound apoM embedded in a POPC bilayer by the N-terminal anchor helix. The region of apoM interacting with the bilayer is highlighted in green.
Figure 6. S1P unbinding process from WT apoM.
The residues located in the hydrophobic lower part are colored wheat. The five residues forming the hydrophilic entrance for S1P are colored yellow. Residues interacting strongly with S1P during enforced unbinding are colored violet. Residues 23–46, 74–83 and 101–126 were omitted in figures (b–e) for clarity.
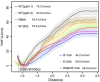
Figure 7. Potential of mean force (PMF) for S1P unbinding.
The PMF was calculated from between 42 and 88 umbrella simulations for each system.
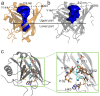
Figure 8. S1P binding pocket of apoM.
(a,b) The binding pockets of S1P-bound apoM (orange) and of ligand-free apoM (grey) are compared to each other. The blue surface borders the measured pocket volumes. The upper pocket, as well as the distance between the C_α_ atoms of residues G93 and Y141 are increased for the ligand-free apoM while the lower part of the cavity is closed. (c) The gating is achieved by Tyr102 and Tyr147 and the pocket occluded by Phe71, Met73, His43, and Ile132 (grey sticks). For bound S1P (cyan sticks), the sidechains (orange sticks) slightly reorient to accomodate the ligand.
Similar articles
- High-Density Lipoprotein-Associated Apolipoprotein M Limits Endothelial Inflammation by Delivering Sphingosine-1-Phosphate to the Sphingosine-1-Phosphate Receptor 1.
Ruiz M, Frej C, Holmér A, Guo LJ, Tran S, Dahlbäck B. Ruiz M, et al. Arterioscler Thromb Vasc Biol. 2017 Jan;37(1):118-129. doi: 10.1161/ATVBAHA.116.308435. Epub 2016 Nov 22. Arterioscler Thromb Vasc Biol. 2017. PMID: 27879252 - Regulation of the metabolism of apolipoprotein M and sphingosine 1-phosphate by hepatic PPARγ activity.
Kurano M, Ikeda H, Iso-O N, Hara M, Tsukamoto K, Yatomi Y. Kurano M, et al. Biochem J. 2018 Jun 21;475(12):2009-2024. doi: 10.1042/BCJ20180052. Biochem J. 2018. PMID: 29712716 - Endothelium-protective sphingosine-1-phosphate provided by HDL-associated apolipoprotein M.
Christoffersen C, Obinata H, Kumaraswamy SB, Galvani S, Ahnström J, Sevvana M, Egerer-Sieber C, Muller YA, Hla T, Nielsen LB, Dahlbäck B. Christoffersen C, et al. Proc Natl Acad Sci U S A. 2011 Jun 7;108(23):9613-8. doi: 10.1073/pnas.1103187108. Epub 2011 May 23. Proc Natl Acad Sci U S A. 2011. PMID: 21606363 Free PMC article. - A Novel Perspective on the ApoM-S1P Axis, Highlighting the Metabolism of ApoM and Its Role in Liver Fibrosis and Neuroinflammation.
Hajny S, Christoffersen C. Hajny S, et al. Int J Mol Sci. 2017 Jul 27;18(8):1636. doi: 10.3390/ijms18081636. Int J Mol Sci. 2017. PMID: 28749426 Free PMC article. Review. - Sphingosine 1-Phosphate and Atherosclerosis.
Kurano M, Yatomi Y. Kurano M, et al. J Atheroscler Thromb. 2018 Jan 1;25(1):16-26. doi: 10.5551/jat.RV17010. Epub 2017 Jul 20. J Atheroscler Thromb. 2018. PMID: 28724841 Free PMC article. Review.
Cited by
- Unbiased MD simulations identify lipid binding sites in lipid transfer proteins.
Srinivasan S, Álvarez D, John Peter AT, Vanni S. Srinivasan S, et al. J Cell Biol. 2024 Nov 4;223(11):e202312055. doi: 10.1083/jcb.202312055. Epub 2024 Aug 6. J Cell Biol. 2024. PMID: 39105757 - The Structural Binding Mode of the Four Autotaxin Inhibitor Types that Differentially Affect Catalytic and Non-Catalytic Functions.
Salgado-Polo F, Perrakis A. Salgado-Polo F, et al. Cancers (Basel). 2019 Oct 16;11(10):1577. doi: 10.3390/cancers11101577. Cancers (Basel). 2019. PMID: 31623219 Free PMC article. Review. - Serum Sphingosine-1-Phosphate Is Decreased in Patients With Acute-on-Chronic Liver Failure and Predicts Early Mortality.
Mücke VT, Maria Schwarzkopf K, Thomas D, Mücke MM, Rüschenbaum S, Trebicka J, Pfeilschifter J, Zeuzem S, Lange CM, Grammatikos G. Mücke VT, et al. Hepatol Commun. 2020 Aug 12;4(10):1477-1486. doi: 10.1002/hep4.1561. eCollection 2020 Oct. Hepatol Commun. 2020. PMID: 33024917 Free PMC article. - A conformation-specific ON-switch for controlling CAR T cells with an orally available drug.
Zajc CU, Dobersberger M, Schaffner I, Mlynek G, Pühringer D, Salzer B, Djinović-Carugo K, Steinberger P, De Sousa Linhares A, Yang NJ, Obinger C, Holter W, Traxlmayr MW, Lehner M. Zajc CU, et al. Proc Natl Acad Sci U S A. 2020 Jun 30;117(26):14926-14935. doi: 10.1073/pnas.1911154117. Epub 2020 Jun 17. Proc Natl Acad Sci U S A. 2020. PMID: 32554495 Free PMC article. - Hepatic FoxOs link insulin signaling with plasma lipoprotein metabolism through an apolipoprotein M/sphingosine-1-phosphate pathway.
Izquierdo MC, Shanmugarajah N, Lee SX, Kraakman MJ, Westerterp M, Kitamoto T, Harris M, Cook JR, Gusarova GA, Zhong K, Marbuary E, O-Sullivan I, Rasmus N, Camastra S, Unterman TG, Ferrannini E, Hurwitz BE, Haeusler RA. Izquierdo MC, et al. J Clin Invest. 2022 Apr 1;132(7):e146219. doi: 10.1172/JCI146219. J Clin Invest. 2022. PMID: 35104242 Free PMC article.
References
- Lee M. J. et al.. Vascular endothelial cell adherens junction assembly and morphogenesis induced by sphingosine-1-phosphate. Cell 99, 301–312 (1999). - PubMed
- Pappu R. et al.. Promotion of lymphocyte egress into blood and lymph by distinct sources of sphingosine-1-phosphate. Science 316, 295–298 (2007). - PubMed
- Prager B., Spampinato S. F. & Ransohoff R. M. Sphingosine 1-phosphate signaling at the blood-brain barrier. Trends Mol. Med. 21, 354–363 (2015). - PubMed
- Spiegel S. & Milstien S. Sphingosine-1-phosphate: signaling inside and out. FEBS Lett. 476, 55–57 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous