Characterization of Outer Membrane Proteome of Akkermansia muciniphila Reveals Sets of Novel Proteins Exposed to the Human Intestine - PubMed (original) (raw)
Characterization of Outer Membrane Proteome of Akkermansia muciniphila Reveals Sets of Novel Proteins Exposed to the Human Intestine
Noora Ottman et al. Front Microbiol. 2016.
Abstract
Akkermansia muciniphila is a common member of the human gut microbiota and belongs to the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum. Decreased levels of A. muciniphila have been associated with many diseases, and thus it is considered to be a beneficial resident of the intestinal mucus layer. Surface-exposed molecules produced by this organism likely play important roles in colonization and communication with other microbes and the host, but the protein composition of the outer membrane (OM) has not been characterized thus far. Herein we set out to identify and characterize A. muciniphila proteins using an integrated approach of proteomics and computational analysis. Sarkosyl extraction and sucrose density-gradient centrifugation methods were used to enrich and fractionate the OM proteome of A. muciniphila. Proteins from these fractions were identified by LC-MS/MS and candidates for OM proteins derived from the experimental approach were subjected to computational screening to verify their location in the cell. In total we identified 79 putative OM and membrane-associated extracellular proteins, and 23 of those were found to differ in abundance between cells of A. muciniphila grown on the natural substrate, mucin, and those grown on the non-mucus sugar, glucose. The identified OM proteins included highly abundant proteins involved in secretion and transport, as well as proteins predicted to take part in formation of the pili-like structures observed in A. muciniphila. The most abundant OM protein was a 95-kD protein, termed PilQ, annotated as a type IV pili secretin and predicted to be involved in the production of pili in A. muciniphila. To verify its location we purified the His-Tag labeled N-terminal domain of PilQ and generated rabbit polyclonal antibodies. Immunoelectron microscopy of thin sections immunolabeled with these antibodies demonstrated the OM localization of PilQ, testifying for its predicted function as a type IV pili secretin in A. muciniphila. As pili structures are known to be involved in the modulation of host immune responses, this provides support for the involvement of OM proteins in the host interaction of A. muciniphila. In conclusion, the characterization of A. muciniphila OM proteome provides valuable information that can be used for further functional and immunological studies.
Keywords: Akkermansia muciniphila; PVC superphylum; gut microbiota; outer membrane; pili; proteomics.
Figures
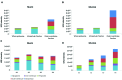
FIGURE 1
Distribution of proteins from different bacterial compartments in isolated fractions. OM fraction isolated from sarkosyl-extracted membranes in comparison to whole proteome and intracellular fraction of both mucin and glucose-grown cells of A. muciniphila (A,B). Fractions of sucrose density-gradient centrifugation (see Supplementary Figure S1) of membranes of A. muciniphila grown on mucin or glucose (C,D). Label-free quantification (LFQ) intensity was used as a proxy for absolute protein abundance. CELLO software was used to predict protein localization.
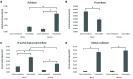
FIGURE 2
Activity of mucin-degrading enzymes in the OM and intracellular fractions. The enzyme activity of sialidase (A), fucosidase (B), _N_-acetyl-β-glucosaminidase (C), and GlcNAc-sulfatase (D) was determined from OM and intracellular (including periplasmic space) fractions of A. muciniphila grown on mucin or glucose as described in Figure 1. Average of duplicate measurements is shown. Significant differences (p < 0.05) in the enzyme activities between samples are indicated with an asterisks.
FIGURE 3
Sarkosyl extraction leads to enrichment of OM and intracellular proteins. Subcellular localization of proteins identified from the OM and intracellular fraction of A. muciniphila grown on mucin (A) or glucose (B). CELLO was used to predict protein localization. Relative abundances of the proteins are presented on a log10 scale with the dashed diagonal line indicating an even distribution between OM and intracellular fraction. A relative abundance of 4.0 represents proteins that were not detected or were under the detection limit. All proteins with LFQ intensity above 8.5 in the sarkosyl-extracted OM fraction (horizontal dashed line) were included in the preliminary list of potential OM proteins.
FIGURE 4
Immunoelectron microscopy labeling of A. muciniphila thin sections visualizes PilQ to be localized on the OM. The cells were labeled with anti-P-Amuc_1098A antibodies and detection was done by using 10-nm protein A-conjugated gold particles. Two representative electronmicrographs are shown.
Similar articles
- Action and function of Akkermansia muciniphila in microbiome ecology, health and disease.
Ottman N, Geerlings SY, Aalvink S, de Vos WM, Belzer C. Ottman N, et al. Best Pract Res Clin Gastroenterol. 2017 Dec;31(6):637-642. doi: 10.1016/j.bpg.2017.10.001. Epub 2017 Oct 13. Best Pract Res Clin Gastroenterol. 2017. PMID: 29566906 Review. - Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function.
Ottman N, Reunanen J, Meijerink M, Pietilä TE, Kainulainen V, Klievink J, Huuskonen L, Aalvink S, Skurnik M, Boeren S, Satokari R, Mercenier A, Palva A, Smidt H, de Vos WM, Belzer C. Ottman N, et al. PLoS One. 2017 Mar 1;12(3):e0173004. doi: 10.1371/journal.pone.0173004. eCollection 2017. PLoS One. 2017. PMID: 28249045 Free PMC article. - Genome-Scale Model and Omics Analysis of Metabolic Capacities of Akkermansia muciniphila Reveal a Preferential Mucin-Degrading Lifestyle.
Ottman N, Davids M, Suarez-Diez M, Boeren S, Schaap PJ, Martins Dos Santos VAP, Smidt H, Belzer C, de Vos WM. Ottman N, et al. Appl Environ Microbiol. 2017 Aug 31;83(18):e01014-17. doi: 10.1128/AEM.01014-17. Print 2017 Sep 15. Appl Environ Microbiol. 2017. PMID: 28687644 Free PMC article. - Nutrient-specific proteomic analysis of the mucin degrading bacterium Akkermansia muciniphila.
Lee JY, Jin HS, Kim KS, Baek JH, Kim BS, Lee DW. Lee JY, et al. Proteomics. 2022 Feb;22(3):e2100125. doi: 10.1002/pmic.202100125. Epub 2021 Oct 10. Proteomics. 2022. PMID: 34596327 - Akkermansia muciniphila in the Human Gastrointestinal Tract: When, Where, and How?
Geerlings SY, Kostopoulos I, de Vos WM, Belzer C. Geerlings SY, et al. Microorganisms. 2018 Jul 23;6(3):75. doi: 10.3390/microorganisms6030075. Microorganisms. 2018. PMID: 30041463 Free PMC article. Review.
Cited by
- Deciphering the trophic interaction between Akkermansia muciniphila and the butyrogenic gut commensal Anaerostipes caccae using a metatranscriptomic approach.
Chia LW, Hornung BVH, Aalvink S, Schaap PJ, de Vos WM, Knol J, Belzer C. Chia LW, et al. Antonie Van Leeuwenhoek. 2018 Jun;111(6):859-873. doi: 10.1007/s10482-018-1040-x. Epub 2018 Feb 19. Antonie Van Leeuwenhoek. 2018. PMID: 29460206 Free PMC article. - Next-Generation Probiotics: Microflora Intervention to Human Diseases.
Zhang H, Duan Y, Cai F, Cao D, Wang L, Qiao Z, Hong Q, Li N, Zheng Y, Su M, Liu Z, Zhu B. Zhang H, et al. Biomed Res Int. 2022 Nov 16;2022:5633403. doi: 10.1155/2022/5633403. eCollection 2022. Biomed Res Int. 2022. PMID: 36440358 Free PMC article. Review. - Akkermansia muciniphila: biology, microbial ecology, host interactions and therapeutic potential.
Ioannou A, Berkhout MD, Geerlings SY, Belzer C. Ioannou A, et al. Nat Rev Microbiol. 2024 Oct 15. doi: 10.1038/s41579-024-01106-1. Online ahead of print. Nat Rev Microbiol. 2024. PMID: 39406893 Review. - Strategies for high cell density cultivation of Akkermansia muciniphila and its potential metabolism.
Wu H, Qi S, Yang R, Pan Q, Lu Y, Yao C, He N, Huang S, Ling X. Wu H, et al. Microbiol Spectr. 2024 Jan 11;12(1):e0238623. doi: 10.1128/spectrum.02386-23. Epub 2023 Dec 7. Microbiol Spectr. 2024. PMID: 38059626 Free PMC article. - Akkermansia muciniphila and Faecalibacterium prausnitzii in Immune-Related Diseases.
Effendi RMRA, Anshory M, Kalim H, Dwiyana RF, Suwarsa O, Pardo LM, Nijsten TEC, Thio HB. Effendi RMRA, et al. Microorganisms. 2022 Nov 30;10(12):2382. doi: 10.3390/microorganisms10122382. Microorganisms. 2022. PMID: 36557635 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials