Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases - PubMed (original) (raw)
Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases
Andrew Janowczyk et al. J Pathol Inform. 2016.
Abstract
Background: Deep learning (DL) is a representation learning approach ideally suited for image analysis challenges in digital pathology (DP). The variety of image analysis tasks in the context of DP includes detection and counting (e.g., mitotic events), segmentation (e.g., nuclei), and tissue classification (e.g., cancerous vs. non-cancerous). Unfortunately, issues with slide preparation, variations in staining and scanning across sites, and vendor platforms, as well as biological variance, such as the presentation of different grades of disease, make these image analysis tasks particularly challenging. Traditional approaches, wherein domain-specific cues are manually identified and developed into task-specific "handcrafted" features, can require extensive tuning to accommodate these variances. However, DL takes a more domain agnostic approach combining both feature discovery and implementation to maximally discriminate between the classes of interest. While DL approaches have performed well in a few DP related image analysis tasks, such as detection and tissue classification, the currently available open source tools and tutorials do not provide guidance on challenges such as (a) selecting appropriate magnification, (b) managing errors in annotations in the training (or learning) dataset, and (c) identifying a suitable training set containing information rich exemplars. These foundational concepts, which are needed to successfully translate the DL paradigm to DP tasks, are non-trivial for (i) DL experts with minimal digital histology experience, and (ii) DP and image processing experts with minimal DL experience, to derive on their own, thus meriting a dedicated tutorial.
Aims: This paper investigates these concepts through seven unique DP tasks as use cases to elucidate techniques needed to produce comparable, and in many cases, superior to results from the state-of-the-art hand-crafted feature-based classification approaches.
Results: Specifically, in this tutorial on DL for DP image analysis, we show how an open source framework (Caffe), with a singular network architecture, can be used to address: (a) nuclei segmentation (F-score of 0.83 across 12,000 nuclei), (b) epithelium segmentation (F-score of 0.84 across 1735 regions), (c) tubule segmentation (F-score of 0.83 from 795 tubules), (d) lymphocyte detection (F-score of 0.90 across 3064 lymphocytes), (e) mitosis detection (F-score of 0.53 across 550 mitotic events), (f) invasive ductal carcinoma detection (F-score of 0.7648 on 50 k testing patches), and (g) lymphoma classification (classification accuracy of 0.97 across 374 images).
Conclusion: This paper represents the largest comprehensive study of DL approaches in DP to date, with over 1200 DP images used during evaluation. The supplemental online material that accompanies this paper consists of step-by-step instructions for the usage of the supplied source code, trained models, and input data.
Keywords: Classification; deep learning; detection; digital histology; machine learning; segmentation.
Figures
Figure 1
The flowchart shows a typical workflow for digital pathology research. Histologic primitives (e.g. nuclei, lymphocytes, mitosis, etc.,) are identified, after which biologically relevant features are extracted for subsequent use in higher order research directives. Typically, the tasks in the red box are undertaken by the development and upkeep of individual task specific approaches. The premise of this tutorial is that these tasks can be performed by a single generic deep learning approach, which can be easily maintained and extended upon
Figure 2
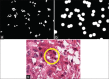
Typical patches extracted for use in training a nuclear segmentation classifier. Six examples of (a) the negative class show large areas of stroma which are notably different than (b) the positive nuclei class and tend to be very easily classified. To compensate, we supplement the training set with (c) patches which are exactly on the edge of the nuclei, forcing the network to learn boundaries better
Figure 3
The process of creation of training exemplars to enhance the result obtained via deep learning for nuclei segmentation. The original image (a) only has (b) a select few of its nuclei annotated. This makes it difficult to find patches which represent a challenging negative class. Our approach involves augmenting a basic negative class, created by sampling from the thresholded color deconvoluted image. More challenging patches are supplied by (c) a dilated edge mask. Sampling locations from (c) allows us to create negative class samples which are of very high utility for the deep learning algorithm. As a result, our improved patch selection technique leads to (e) notably better-delineated nuclei boundaries as compared to the approach shown in (d)
Figure 4
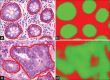
Nuclear segmentation output as produced by our approach wherein the original image in (a) is shown with (b) the associated manually annotated ground truth. When applying the network at × 40 probability map (c) is obtained, where the more green a pixel is, the higher the probability associated with it belonging to the nuclei class. The × 20 version is shown in (d)
Figure 5
Epithelium segmentation output as produced by our approach where original images in (a and d) have their associated ground truth in (b and e) overlaid. We can see that the results from the deep learning, in (c and f), that a pixel level metric is perhaps not ultimately suited to quantify this task as deep learning is better able to provide a pixel level classification, intractable for a human expert to parallel
Figure 6
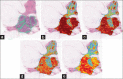
The benign tubules, outlined in red, (a) are more organized and similar, as a result the deep learning can provide very clear boundaries (b), where the stronger green indicates a higher likelihood that a pixel belongs to the tubule class. On the other hand, when considering malignant tubules (c), the variances are quite large making it more difficult for a learn from data approach to generalize to all unseen cases. Our results (d) are able to identify a large portion of the associated pixels, but can be seen providing incorrect labeling in situations where traditional structures are not present
Figure 7
Invasive ductal carcinoma segmentation where we see the original sample (a) with the pathologist annotated region shown in green. From (b) we can see the results generated by the resizing approach, (c) shows the same results without resizing, (d) shows the output when resizing and balancing the training set and (e) finally resizing with dropout, where the more red a pixel is, the more likely it represents an invasive ductal carcinomas pixel. We note that the upper half of the image actually contains true positives which were not annotated by the pathologist
Figure 8
Lymphocyte detection result where green dots are the ground truth, and red dots are the centers discovered by the algorithm. The image on the left (a) has 21 TP/2 FP/0 FN. The false positives are on the edges, about 1 o’clock and 3 o’clock. The image on the right (b) one has 11 TP/1 FP/2 FN. We can see the false negatives are quite small and not very clear making it hard to detect them without also encountering many false positives. The only false positive is in the middle at around 7 o’clock though this structure does look “lymphocyte-like
Figure 9
Result of deep learning for mitosis detection, where the blue ratio segmentation approach is used to generate the initial result in (a). We take this input and dilate it to greatly reduce the total area of interest in a sample. (b) In the final image, (c) we can see that the mitosis is indeed located in the middle of the image, included our computational mask. We can see that the mitosis is in the telophase stage, such that the DNA components have split into two pieces (in yellow circle), making it more difficult to identify
Figure 10
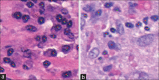
False positive samples of mitoses (a) with (b) true positive samples on the right. We can see that in many cases the two classes are indistinguishable from each other in the two-dimensional plane, thus requiring the common practice of focal length manipulation of the microscope to determine which instances are truly mitotic events
Figure 11
Exemplars taken from the (a) chronic lymphocytic leukemia, (b) follicular lymphoma, and (c) mantle cell lymphoma classes used in this task. There is notable staining difference across the three samples. Also, it is not intuitively obvious what the characteristics are which should be used to classify these images
Figure 12
(a and b) Misclassified image belonging to the follicular lymphoma subtype. We can see that when magnified, there appears to be some type of artifact created during the scanning process. It is not unreasonable to think that upon seeing this a clinician would ask for it to be rescanned
Similar articles
- The impact of pre- and post-image processing techniques on deep learning frameworks: A comprehensive review for digital pathology image analysis.
Salvi M, Acharya UR, Molinari F, Meiburger KM. Salvi M, et al. Comput Biol Med. 2021 Jan;128:104129. doi: 10.1016/j.compbiomed.2020.104129. Epub 2020 Nov 21. Comput Biol Med. 2021. PMID: 33254082 Review. - Deep learning in digital pathology image analysis: a survey.
Deng S, Zhang X, Yan W, Chang EI, Fan Y, Lai M, Xu Y. Deng S, et al. Front Med. 2020 Aug;14(4):470-487. doi: 10.1007/s11684-020-0782-9. Epub 2020 Jul 29. Front Med. 2020. PMID: 32728875 Review. - Deep computational pathology in breast cancer.
Duggento A, Conti A, Mauriello A, Guerrisi M, Toschi N. Duggento A, et al. Semin Cancer Biol. 2021 Jul;72:226-237. doi: 10.1016/j.semcancer.2020.08.006. Epub 2020 Aug 17. Semin Cancer Biol. 2021. PMID: 32818626 Review. - A resolution adaptive deep hierarchical (RADHicaL) learning scheme applied to nuclear segmentation of digital pathology images.
Janowczyk A, Doyle S, Gilmore H, Madabhushi A. Janowczyk A, et al. Comput Methods Biomech Biomed Eng Imaging Vis. 2018;6(3):270-276. doi: 10.1080/21681163.2016.1141063. Epub 2016 Apr 28. Comput Methods Biomech Biomed Eng Imaging Vis. 2018. PMID: 29732269 Free PMC article. - Quantitative Assessment of the Effects of Compression on Deep Learning in Digital Pathology Image Analysis.
Chen Y, Janowczyk A, Madabhushi A. Chen Y, et al. JCO Clin Cancer Inform. 2020 Mar;4:221-233. doi: 10.1200/CCI.19.00068. JCO Clin Cancer Inform. 2020. PMID: 32155093 Free PMC article.
Cited by
- A Deep Multi-Label Segmentation Network For Eosinophilic Esophagitis Whole Slide Biopsy Diagnostics.
Daniel N, Larey A, Aknin E, Osswald GA, Caldwell JM, Rochman M, Collins MH, Yang GY, Arva NC, Capocelli KE, Rothenberg ME, Savir Y. Daniel N, et al. Annu Int Conf IEEE Eng Med Biol Soc. 2022 Jul;2022:3211-3217. doi: 10.1109/EMBC48229.2022.9871086. Annu Int Conf IEEE Eng Med Biol Soc. 2022. PMID: 36085661 Free PMC article. - DFUCare: deep learning platform for diabetic foot ulcer detection, analysis, and monitoring.
Sendilraj V, Pilcher W, Choi D, Bhasin A, Bhadada A, Bhadadaa SK, Bhasin M. Sendilraj V, et al. Front Endocrinol (Lausanne). 2024 Sep 23;15:1386613. doi: 10.3389/fendo.2024.1386613. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39381435 Free PMC article. - Artificial intelligence neuropathologist for glioma classification using deep learning on hematoxylin and eosin stained slide images and molecular markers.
Jin L, Shi F, Chun Q, Chen H, Ma Y, Wu S, Hameed NUF, Mei C, Lu J, Zhang J, Aibaidula A, Shen D, Wu J. Jin L, et al. Neuro Oncol. 2021 Jan 30;23(1):44-52. doi: 10.1093/neuonc/noaa163. Neuro Oncol. 2021. PMID: 32663285 Free PMC article. - Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology.
Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Bera K, et al. Nat Rev Clin Oncol. 2019 Nov;16(11):703-715. doi: 10.1038/s41571-019-0252-y. Epub 2019 Aug 9. Nat Rev Clin Oncol. 2019. PMID: 31399699 Free PMC article. Review.
References
- Veta M, Pluim JP, van Diest PJ, Viergever MA. Breast cancer histopathology image analysis: A review. IEEE Trans Biomed Eng. 2014;61:1400–11. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources