Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism - PubMed (original) (raw)
. 2016 Sep 8;166(6):1572-1584.e16.
doi: 10.1016/j.cell.2016.08.006. Epub 2016 Sep 1.
Christoph A Weber 2, Marco Nousch 3, Omar Adame-Arana 2, Carsten Hoege 1, Marco Y Hein 4, Erin Osborne-Nishimura 5, Julia Mahamid 4, Marcus Jahnel 1, Louise Jawerth 6, Andrej Pozniakovski 1, Christian R Eckmann 3, Frank Jülicher 7, Anthony A Hyman 8
Affiliations
- PMID: 27594427
- PMCID: PMC5034880
- DOI: 10.1016/j.cell.2016.08.006
Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism
Shambaditya Saha et al. Cell. 2016.
Abstract
P granules are non-membrane-bound RNA-protein compartments that are involved in germline development in C. elegans. They are liquids that condense at one end of the embryo by localized phase separation, driven by gradients of polarity proteins such as the mRNA-binding protein MEX-5. To probe how polarity proteins regulate phase separation, we combined biochemistry and theoretical modeling. We reconstitute P granule-like droplets in vitro using a single protein PGL-3. By combining in vitro reconstitution with measurements of intracellular concentrations, we show that competition between PGL-3 and MEX-5 for mRNA can regulate the formation of PGL-3 droplets. Using theory, we show that, in a MEX-5 gradient, this mRNA competition mechanism can drive a gradient of P granule assembly with similar spatial and temporal characteristics to P granule assembly in vivo. We conclude that gradients of polarity proteins can position RNP granules during development by using RNA competition to regulate local phase separation.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
Figure 1
PGL-3 forms liquid-like drops in vitro. (A) SDS-PAGE of PGL-3-mEGFP expressed and purified from insect cells. (B) Maximum intensity projection of series of confocal z- slices shows PGL-3-mEGFP at 10 μM phase-separates into drops. (C) Virtual slice from cryo-electron tomography, 5 nm thick, shows a drop of PGL-3-mEGFP (1 μM). Red and blue boxes show zoomed in view. (D) Time-lapse single confocal plane micrographs show two PGL-3-mEGFP drops fuse with each other into a single drop within 3.5 s. (E) Time-lapse single confocal plane micrographs show fluorescence recovery of PGL-3-mEGFP after an internal 1.44 μm × 1.44 μm area is photobleached at 3 s. (F) Quantification of FRAP data presented in (E), n = 28, error bars represent 1 SD. See also Figure S1, Movies S1–S2.
Figure 2
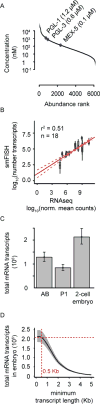
Measurements of intracellular protein and mRNA concentration in C. elegans embryo. (A) Plot of the concentration and abundance of different proteins measured by mass spectrometry in C. elegans early-embryo extracts. (B) Correlation plot of RNA-seq intensity values and corresponding smFISH single molecule counts using a dataset of 9 genes in both blastomeres (AB and P1 cells; 18 samples total) (Osborne Nishimura, PLoS Genetics 2015). Red line: Fit using linear regression analysis. Dotted red lines: 95% confidence interval. (C) Estimates of total mRNA transcripts per blastomere (AB and P1 cells) and for the entire 2-cell embryo are shown. Error bars represent 95% confidence intervals. (D) Cumulative frequency distribution for minimum lengths of mRNA transcripts in the 2-cell stage embryo (AB and P1 total). Grey shaded areas represent 95% confidence intervals. Red broken lines highlight the total number of mRNA transcripts predicted if only mRNA transcripts longer than 500 bases are included in the estimate. See also Figure S2, Tables S1–S2.
Figure 3
Messenger RNA promotes PGL-3 drop assembly depending on RGG repeats in PGL-3. (A) Cartoon describing wild-type PGL-3 (WT) and the construct RGG_mut. RGG_mut: PGL-3 construct with arginine in all six RGG repeats (yellow boxes) mutated to either glycine or leucine. (B) Maximum intensity projection of series of confocal z- slices show drop formation of mEGFP-tagged PGL-3 or RGG_mut (0.6 μM) in presence or absence of 10 ng/μL total C. elegans mRNA. Red box shows zoomed in view of drops. (C) Quantification of data presented in (B). In each case, 20 observation volumes (41 μm × 41 μm × ~10 μm) were scored. Number of drops observed in each observation volume is represented as a grey circle, or triangle in the plot. The mean is shown in red. Error bars are 1 SEM. (D) Plot of the fraction of total GFP fluorescence found in phase-separated drops as a function of total PGL-3-mEGFP concentration in presence or absence of 50 ng/μL mouse brain mRNA. For each concentration of PGL-3, drops in ≥12 observation volumes (41 μm × 41 μm × ~10 μm) were scored. Error bars represent 1 SEM among the observation volumes scored. (E–G) Quantification of the number of PGL-3-mEGFP drops scored under different conditions. In each case, 20 observation volumes (E, G: 41 μm × 41 μm × ~10 μm; F: 71 μm × 71 μm × ~10 μm) were scored. Number of drops observed in each observation volume is represented as a grey triangle, circle, diamond, or square in the plot. The mean is shown in red. Error bars are 1 SD. (E) Drop assembly on addition of different kinds of RNA to PGL-3-mEGFP (0.6 μM). Triangles: no RNA, circles: total C. elegans mRNA (10 ng/μL), or in vitro transcribed luciferase mRNA lacking 5′ cap and poly(A) tail (10 ng/μL), diamonds: in vitro transcribed 18S C. elegans rRNA (10 ng/μL), squares: total RNA from C. elegans (10 or 200 ng/μL). (F) Drop assembly on addition of in vitro transcribed 18S rRNA pre-heated at 75 °C for 1 min to PGL-3-mEGFP (0.6 μM). Circles: no RNA (control buffer pre-heated), triangles: 18S rRNA (40 nM) with or without pre-heating. (G) Drop assembly on addition of fragments of in vitro transcribed luciferase mRNA lacking 5′ cap and poly(A) tail to PGL-3-mEGFP (0.6 μM). Circles: no RNA, triangles: luciferase mRNA fragments 200, 400, 600 or 800 bases long (20 nM). (H) Maximum intensity projection of series of confocal z- slices show colocalization of PGL-3-mEGFP (0.25 μM) and cyanine-5 labeled luciferase mRNA (3 ng/μL) in drops. See also Figures S1, S3, S6.
Figure 4
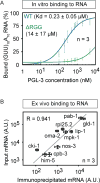
MEX-5 inhibits mRNA-dependent PGL-3 drop assembly. (A) Maximum intensity projections of series of confocal z- slices show PGL-3-mEGFP (0.6 μM) in presence or absence of additives: 1) PGL-3 alone, 2) + 150 nM MBP-MEX-5 (236–350), 3) + 50 ng/μL mouse brain mRNA, 4) + 50 ng/μL mouse brain mRNA and 150 nM MBP-MEX-5 (236–350). (B) Plot of the fraction of total GFP fluorescence found in phase-separated drops of PGL-3-mEGFP (0.6 μM) in presence of different additives: 1) + 150 nM MBP-MEX-5 (236–350), 2) + total mouse brain mRNA (50 ng/μL), and 3–5) + MBP-MEX-5 (236–350) (150 nM) and total mouse brain mRNA (50, 100, or 150 ng/μL). In each case, drops in ≥16 observation volumes (41 μm × 41 μm × ~10 μm) were scored. Error bars represent 1 SEM among the observation volumes scored. (C) Binding of MBP-MEX-5 (236–350) to RNA in vitro in filter binding assay. Plot shows the amount of (GUU)10A10 RNA oligo bound to MBP-MEX-5 (236–350) as a function of protein concentration. Error bars represent 1 SEM. (D) Binding of MBP-MEX-5 (236–350) to PGL-3-mEGFP in vitro in pull-down assay using beads coated with anti-PGL-3 antibody. Plot of the fraction of MBP-MEX-5 (236–350) or PGL-3-mEGFP present in supernatant and pellet. Control: MBP-MEX-5 (236–350) alone (1 μM), Experiment: MBP-MEX-5 (236–350) (1 μM) and PGL-3-mEGFP (1 μM). Error bars represent 1 SD among three independent experiments. See also Figures S4, S6.
Figure 5
PGL-3 binds weakly to mRNA with low sequence specificity. (A) Binding of PGL-3 to RNA in vitro in filter binding assay. Plot shows the amount of (GUU)10A10 RNA oligo bound to PGL-3-mEGFP (blue diamonds) or ΔRGG-mEGFP (green diamonds) as a function of protein concentration. Error bars represent 1 SEM. ΔRGG is a PGL-3 construct that lacks the C-terminal 60 residues in PGL-3 containing the RGG repeats. (B) Binding of PGL-3 to RNA ex vivo assayed in co-immunoprecipitation of RNA with PGL-3-mEGFP from a pool of total RNA purified from C. elegans germline. Correlation plot of the amount of ten different mRNA species in input and the amount co-IPed with PGL-3-mEGFP. R is Pearson coefficient. See also Figure S5.
Figure 6
Theoretical model of PGL-3 phase separation, mRNA binding and interactions with MEX-5. (A) Experimentally determined difference ΔI between concentrations of PGL-3 inside and outside of drops for different overall PGL-3 concentration (cPT) in the presence (red dots) or in the absence (blue squares) of 50 ng/μL mouse brain mRNA. This is a different representation of the data presented in Figure 3D. For details on measurement of ΔI, please see methods section. Solid lines are the corresponding fits obtained from our theoretical model. Vertical arrows indicate the threshold concentrations above which phase separated droplets form in presence of mRNA (cPR*) or in absence of mRNA (cP*). Error bars represent 1 SEM. (B) Phase diagram calculated from our model for the ternary mixture consisting of PGL-3, PGL-3 bound to mRNA (PGL-3:mRNA) and water. As the total concentration of PGL-3 (cPT) is increased keeping the total concentration of mRNA constant (along the orange line in the phase diagram), mRNA binds to PGL-3 and the system equilibrates to certain concentrations of PGL-3 (cP) and PGL-3:mRNA (cPR). The binodal lines (purple) split the regions where the solution is mixed (no drops form) or demixed (drops form via phase separation). In absence of drops in the mixed region of the phase diagram, the system equilibrates to a unique concentration of PGL-3 (cP) and PGL-3:mRNA (cPR). In the drop-containing demixed region of the phase diagram, for a given total concentration of PGL-3 and mRNA, there are two distinct sets of values of cP and cPR corresponding to concentrations inside and outside of drops. The green lines connecting these two sets of concentration values are called ‘tie lines’. Using the tie lines, we can construct the behavior of ΔI as a function of cPT as shown in A (for more details see methods section and movie S3). (C–D) Results from numerical calculations for the six-component system consisting of mRNA, PGL-3, PGL-3:mRNA, MEX-5, MEX-5:mRNA and water. (C) Snapshots of the total PGL-3 concentration as a function of time and space (left) and plots at each time point of the concentration of total MEX-5 and total mRNA (averaged over the y-coordinate) as a function of position along x-axis (right). (D) Representative time series show dissolution of a drop in a region of high MEX-5 concentration. mRNA is depleted from drops before the drop dissolves. See also Figure S6, Movies S3–S5, Table S3.
Figure 7
Model mechanism of inhibition of mRNA-dependent PGL-3 drop assembly by MEX-5. In absence of MEX-5, mRNA binds PGL-3 via the RGG repeats and increases the local concentration of PGL-3, leading to phase separation. Concentration of mRNA and PGL-3 is significantly higher in the drop phase compared to the surrounding bulk phase. In presence of MEX-5, mRNA binds preferably to MEX-5 in contrast to PGL-3 resulting in inhibition of drop assembly. mRNA molecules not bound to MEX-5 bind PGL-3 and assemble few drops. These drops may recruit few mRNA-MEX-5 complexes.
Similar articles
- Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3.
Smith J, Calidas D, Schmidt H, Lu T, Rasoloson D, Seydoux G. Smith J, et al. Elife. 2016 Dec 3;5:e21337. doi: 10.7554/eLife.21337. Elife. 2016. PMID: 27914198 Free PMC article. - A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos.
Putnam A, Cassani M, Smith J, Seydoux G. Putnam A, et al. Nat Struct Mol Biol. 2019 Mar;26(3):220-226. doi: 10.1038/s41594-019-0193-2. Epub 2019 Mar 4. Nat Struct Mol Biol. 2019. PMID: 30833787 Free PMC article. - C. elegans germ granules require both assembly and localized regulators for mRNA repression.
Aoki ST, Lynch TR, Crittenden SL, Bingman CA, Wickens M, Kimble J. Aoki ST, et al. Nat Commun. 2021 Feb 12;12(1):996. doi: 10.1038/s41467-021-21278-1. Nat Commun. 2021. PMID: 33579952 Free PMC article. - The P Granules of C. elegans: A Genetic Model for the Study of RNA-Protein Condensates.
Seydoux G. Seydoux G. J Mol Biol. 2018 Nov 2;430(23):4702-4710. doi: 10.1016/j.jmb.2018.08.007. Epub 2018 Aug 8. J Mol Biol. 2018. PMID: 30096346 Free PMC article. Review. - Acto-myosin reorganization and PAR polarity in C. elegans.
Cowan CR, Hyman AA. Cowan CR, et al. Development. 2007 Mar;134(6):1035-43. doi: 10.1242/dev.000513. Epub 2007 Feb 7. Development. 2007. PMID: 17287245 Review.
Cited by
- RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates.
Garcia-Jove Navarro M, Kashida S, Chouaib R, Souquere S, Pierron G, Weil D, Gueroui Z. Garcia-Jove Navarro M, et al. Nat Commun. 2019 Jul 19;10(1):3230. doi: 10.1038/s41467-019-11241-6. Nat Commun. 2019. PMID: 31324804 Free PMC article. - Three archetypical classes of macromolecular regulators of protein liquid-liquid phase separation.
Ghosh A, Mazarakos K, Zhou HX. Ghosh A, et al. Proc Natl Acad Sci U S A. 2019 Sep 24;116(39):19474-19483. doi: 10.1073/pnas.1907849116. Epub 2019 Sep 10. Proc Natl Acad Sci U S A. 2019. PMID: 31506351 Free PMC article. - Formation of biological condensates via phase separation: Characteristics, analytical methods, and physiological implications.
Feng Z, Chen X, Wu X, Zhang M. Feng Z, et al. J Biol Chem. 2019 Oct 4;294(40):14823-14835. doi: 10.1074/jbc.REV119.007895. Epub 2019 Aug 23. J Biol Chem. 2019. PMID: 31444270 Free PMC article. Review. - Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation.
Tsang B, Arsenault J, Vernon RM, Lin H, Sonenberg N, Wang LY, Bah A, Forman-Kay JD. Tsang B, et al. Proc Natl Acad Sci U S A. 2019 Mar 5;116(10):4218-4227. doi: 10.1073/pnas.1814385116. Epub 2019 Feb 14. Proc Natl Acad Sci U S A. 2019. PMID: 30765518 Free PMC article. - Switching states: dynamic remodelling of polarity complexes as a toolkit for cell polarization.
Peglion F, Goehring NW. Peglion F, et al. Curr Opin Cell Biol. 2019 Oct;60:121-130. doi: 10.1016/j.ceb.2019.05.002. Epub 2019 Jul 9. Curr Opin Cell Biol. 2019. PMID: 31295650 Free PMC article. Review.
References
- Barber CB, Dobkin DP, Huhdanpaa H. The quickhull algorithm for convex hulls. ACM Transactions on Mathematical Software (TOMS) 1996;22:469–483.
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. - PubMed
- Bray AJ. Theory of phase-ordering kinetics. Advances in Physics. 1994;43:357–459.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources