Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense - PubMed (original) (raw)
Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense
Vera Bunesova et al. BMC Microbiol. 2016.
Abstract
Background: Human milk oligosaccharides (HMOs) are one of the major glycan source of the infant gut microbiota. The two species that predominate the infant bifidobacteria community, Bifidobacterium longum subsp. infantis and Bifidobacterium bifidum, possess an arsenal of enzymes including α-fucosidases, sialidases, and β-galactosidases to metabolise HMOs. Recently bifidobacteria were obtained from the stool of six month old Kenyan infants including species such as Bifidobacterium kashiwanohense, and Bifidobacterium pseudolongum that are not frequently isolated from infant stool. The aim of this study was to characterize HMOs utilization by these isolates. Strains were grown in presence of 2'-fucosyllactose (2'-FL), 3'-fucosyllactose (3'-FL), 3'-sialyl-lactose (3'-SL), 6'-sialyl-lactose (6'-SL), and Lacto-N-neotetraose (LNnT). We further investigated metabolites formed during L-fucose and fucosyllactose utilization, and aimed to identify genes and pathways involved through genome comparison.
Results: Bifidobacterium longum subsp. infantis isolates, Bifidobacterium longum subsp. suis BSM11-5 and B. kashiwanohense strains grew in the presence of 2'-FL and 3'- FL. All B. longum isolates utilized the L-fucose moiety, while B. kashiwanohense accumulated L-fucose in the supernatant. 1,2-propanediol (1,2-PD) was the major metabolite from L-fucose fermentation, and was formed in equimolar amounts by B. longum isolates. Alpha-fucosidases were detected in all strains that degraded fucosyllactose. B. longum subsp. infantis TPY11-2 harboured four α-fucosidases with 95-99 % similarity to the type strain. B. kashiwanohense DSM 21854 and PV20-2 possessed three and one α-fucosidase, respectively. The two α-fucosidases of B. longum subsp. suis were 78-80 % similar to B. longum subsp. infantis and were highly similar to B. kashiwanohense α-fucosidases (95-99 %). The genomes of B. longum strains that were capable of utilizing L-fucose harboured two gene regions that encoded enzymes predicted to metabolize L-fucose to L-lactaldehyde, the precursor of 1,2-PD, via non-phosphorylated intermediates.
Conclusion: Here we observed that the ability to utilize fucosyllactose is a trait of various bifidobacteria species. For the first time, strains of B. longum subsp. infantis and an isolate of B. longum subsp. suis were shown to use L-fucose to form 1,2-PD. As 1,2-PD is a precursor for intestinal propionate formation, bifidobacterial L-fucose utilization may impact intestinal short chain fatty acid balance. A L-fucose utilization pathway for bifidobacteria is suggested.
Keywords: 1,2 propanediol; Bifidobacterium; HMOs; L-fucose; fucosyllactose.
Figures
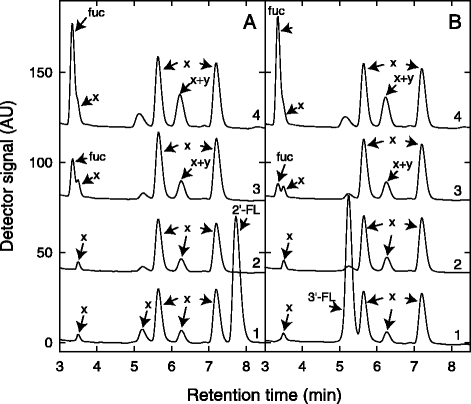
Fig. 1
Degradation of 2′-FL (a) and 3′-FL (b) and accumulation of L-fucose. Shown are (1) unfermented control, (2) B. longum subsp. infantis DSM 20088, (3) B. kashiwanohense PV20-2, and (4) B. bifidum BSM28-1 as representatives of B. longum, B. kashiwanohense and B. bifidum isolates investigated. x, undefined media components; y, intermediate degradation compound of fucosyllactose metabolism; fuc, L-fucose
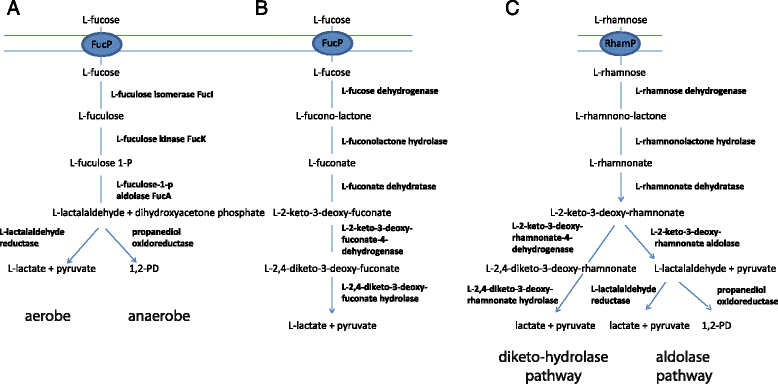
Fig. 2
Comparison of L-fucose (a, b) and L-rhamnose (c) dedegradation pathways in E. coli (a), X. campestris (b), and Sphingomonas sp. (c) extracted from Boronat and Aguilar [30], Yew et al. [44], and Watanabe and Makino [45]
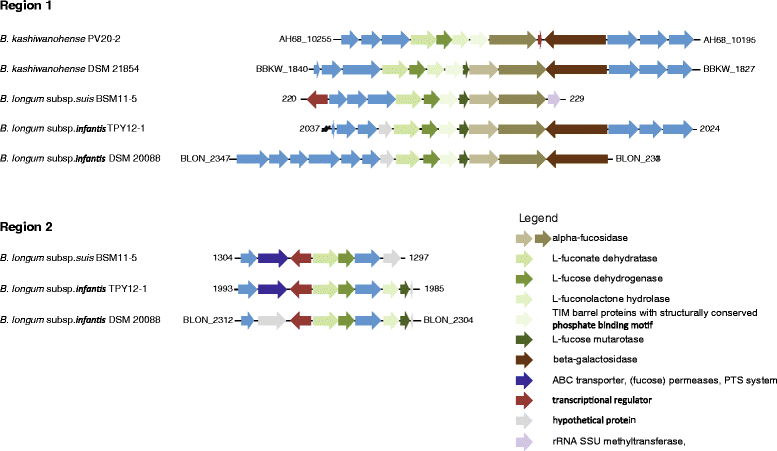
Fig. 3
Genomic regions encompassing genes putatively involved in fucose degradation in B. longum and B. kashiwanohense strains. The gene cluster of region 1 also contained genes encoding the α-fucosidases BLON_2335 and BLON_2336 and is part of the B. longum subsp. infantis HMO utilization operon H1 [3, 11] (not drawn according to scale)
Similar articles
- Trophic Interactions of Infant Bifidobacteria and Eubacterium hallii during L-Fucose and Fucosyllactose Degradation.
Schwab C, Ruscheweyh HJ, Bunesova V, Pham VT, Beerenwinkel N, Lacroix C. Schwab C, et al. Front Microbiol. 2017 Jan 30;8:95. doi: 10.3389/fmicb.2017.00095. eCollection 2017. Front Microbiol. 2017. PMID: 28194144 Free PMC article. - Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization.
LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. LoCascio RG, et al. Appl Environ Microbiol. 2010 Nov;76(22):7373-81. doi: 10.1128/AEM.00675-10. Epub 2010 Aug 27. Appl Environ Microbiol. 2010. PMID: 20802066 Free PMC article. - Novel Genes and Metabolite Trends in Bifidobacterium longum subsp. infantis Bi-26 Metabolism of Human Milk Oligosaccharide 2'-fucosyllactose.
Zabel B, Yde CC, Roos P, Marcussen J, Jensen HM, Salli K, Hirvonen J, Ouwehand AC, Morovic W. Zabel B, et al. Sci Rep. 2019 May 28;9(1):7983. doi: 10.1038/s41598-019-43780-9. Sci Rep. 2019. PMID: 31138818 Free PMC article. - Varied Pathways of Infant Gut-Associated Bifidobacterium to Assimilate Human Milk Oligosaccharides: Prevalence of the Gene Set and Its Correlation with Bifidobacteria-Rich Microbiota Formation.
Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao JZ, Kitaoka M, Katayama T. Sakanaka M, et al. Nutrients. 2019 Dec 26;12(1):71. doi: 10.3390/nu12010071. Nutrients. 2019. PMID: 31888048 Free PMC article. Review. - Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides.
Sela DA, Mills DA. Sela DA, et al. Trends Microbiol. 2010 Jul;18(7):298-307. doi: 10.1016/j.tim.2010.03.008. Epub 2010 Apr 19. Trends Microbiol. 2010. PMID: 20409714 Free PMC article. Review.
Cited by
- Impact of 2'-Fucosyllactose on Gut Microbiota Composition in Adults with Chronic Gastrointestinal Conditions: Batch Culture Fermentation Model and Pilot Clinical Trial Findings.
Ryan JJ, Monteagudo-Mera A, Contractor N, Gibson GR. Ryan JJ, et al. Nutrients. 2021 Mar 14;13(3):938. doi: 10.3390/nu13030938. Nutrients. 2021. PMID: 33799455 Free PMC article. - _Bifidobacterium longum_-fermented rice bran and rice bran supplementation affects the gut microbiome and metabolome.
Nealon NJ, Parker KD, Lahaie P, Ibrahim H, Maurya AK, Raina K, Ryan EP. Nealon NJ, et al. Benef Microbes. 2019 Dec 9;10(8):823-839. doi: 10.3920/BM2019.0017. Epub 2019 Sep 29. Benef Microbes. 2019. PMID: 31965839 Free PMC article. - The genus bifidobacterium: From genomics to functionality of an important component of the mammalian gut microbiota running title: Bifidobacterial adaptation to and interaction with the host.
Alessandri G, van Sinderen D, Ventura M. Alessandri G, et al. Comput Struct Biotechnol J. 2021 Mar 9;19:1472-1487. doi: 10.1016/j.csbj.2021.03.006. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33777340 Free PMC article. Review. - Fecal microbiome and metabolome of infants fed bovine MFGM supplemented formula or standard formula with breast-fed infants as reference: a randomized controlled trial.
He X, Parenti M, Grip T, Lönnerdal B, Timby N, Domellöf M, Hernell O, Slupsky CM. He X, et al. Sci Rep. 2019 Aug 12;9(1):11589. doi: 10.1038/s41598-019-47953-4. Sci Rep. 2019. PMID: 31406230 Free PMC article. Clinical Trial. - The Role of Two Human Milk Oligosaccharides, 2'-Fucosyllactose and Lacto-N-Neotetraose, in Infant Nutrition.
Hegar B, Wibowo Y, Basrowi RW, Ranuh RG, Sudarmo SM, Munasir Z, Atthiyah AF, Widodo AD, Supriatmo, Kadim M, Suryawan A, Diana NR, Manoppo C, Vandenplas Y. Hegar B, et al. Pediatr Gastroenterol Hepatol Nutr. 2019 Jul;22(4):330-340. doi: 10.5223/pghn.2019.22.4.330. Epub 2019 Jun 25. Pediatr Gastroenterol Hepatol Nutr. 2019. PMID: 31338308 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials