Budding Yeast: An Ideal Backdrop for In vivo Lipid Biochemistry - PubMed (original) (raw)
Review
Budding Yeast: An Ideal Backdrop for In vivo Lipid Biochemistry
Pushpendra Singh. Front Cell Dev Biol. 2017.
Abstract
Biological membranes are non-covalent assembly of lipids and proteins. Lipids play critical role in determining membrane physical properties and regulate the function of membrane associated proteins. Budding yeast Saccharomyces cerevisiae offers an exceptional advantage to understand the lipid-protein interactions since lipid metabolism and homeostasis are relatively simple and well characterized as compared to other eukaryotes. In addition, a vast array of genetic and cell biological tools are available to determine and understand the role of a particular lipid in various lipid metabolic disorders. Budding yeast has been instrumental in delineating mechanisms related to lipid metabolism, trafficking and their localization in different subcellular compartments at various cell cycle stages. Further, availability of tools and enormous potential for the development of useful reagents and novel technologies to localize a particular lipid in different subcellular compartments in yeast makes it a formidable system to carry out lipid biology. Taken together, yeast provides an outstanding backdrop to characterize lipid metabolic changes under various physiological conditions.
Keywords: budding yeast; ergosterol; lipid sensors; lipid-protein interactions; sphingolipid.
Figures
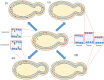
Figure 1
Scheme depicting different lipid modulations achieved in budding yeast. A thick cell wall is present around the plasma membrane carrying proteins (shown in cyan, orange, and yellow just to distinguish them as different) in budding yeast. Cell wall and lipids are depicted in orange and blue color, respectively. A handful of examples are presented (i) visualizing lipids with development of fluorescent biosensors as red star (Fairn et al., ; Das et al., 2012) (ii) remodeling membrane to be thicker by acyl chain lengthening (Dickson et al., ; Gaigg et al., ; Renne et al., 2015) (iii) producing thinner membrane by acyl chain shortening (Cerantola et al., ; Epstein et al., ; Renne et al., 2015) (iv) converting ergosterol to cholesterol in budding yeast (Souza et al., 2011). See text for more details.
Similar articles
- The COP9 signalosome is involved in the regulation of lipid metabolism and of transition metals uptake in Saccharomyces cerevisiae.
Licursi V, Salvi C, De Cesare V, Rinaldi T, Mattei B, Fabbri C, Serino G, Bramasole L, Zimbler JZ, Pick E, Barnes BM, Bard M, Negri R. Licursi V, et al. FEBS J. 2014 Jan;281(1):175-90. doi: 10.1111/febs.12584. Epub 2013 Nov 25. FEBS J. 2014. PMID: 24164706 - Yeast membrane lipid imbalance leads to trafficking defects toward the Golgi.
Woodman S, Trousdale C, Conover J, Kim K. Woodman S, et al. Cell Biol Int. 2018 Jul;42(7):890-902. doi: 10.1002/cbin.10956. Epub 2018 Apr 19. Cell Biol Int. 2018. PMID: 29500884 - Lipid droplet-mediated lipid and protein homeostasis in budding yeast.
Graef M. Graef M. FEBS Lett. 2018 Apr;592(8):1291-1303. doi: 10.1002/1873-3468.12996. Epub 2018 Feb 16. FEBS Lett. 2018. PMID: 29397034 Free PMC article. Review. - Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae.
Bagnat M, Simons K. Bagnat M, et al. Biol Chem. 2002 Oct;383(10):1475-80. doi: 10.1515/BC.2002.169. Biol Chem. 2002. PMID: 12452424 - Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae.
Daum G, Lees ND, Bard M, Dickson R. Daum G, et al. Yeast. 1998 Dec;14(16):1471-510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. Yeast. 1998. PMID: 9885152 Review.
Cited by
- Regulation of yeast polarized exocytosis by phosphoinositide lipids.
Volpiana MW, Nenadic A, Beh CT. Volpiana MW, et al. Cell Mol Life Sci. 2024 Nov 19;81(1):457. doi: 10.1007/s00018-024-05483-x. Cell Mol Life Sci. 2024. PMID: 39560727 Review. - Harnessing Clickable Acylated Glycerol Probes as Chemical Tools for Tracking Glycerolipid Metabolism.
Ancajas CF, Carr AJ, Lou J, Sagar R, Zhou Y, Reynolds TB, Best MD. Ancajas CF, et al. Chemistry. 2023 Jul 6;29(38):e202300417. doi: 10.1002/chem.202300417. Epub 2023 May 15. Chemistry. 2023. PMID: 37085958 Free PMC article. - Many ways, one microorganism: Several approaches to study Malassezia in interactions with model hosts.
Ehemann K, Mantilla MJ, Mora-Restrepo F, Rios-Navarro A, Torres M, Celis Ramírez AM. Ehemann K, et al. PLoS Pathog. 2022 Sep 8;18(9):e1010784. doi: 10.1371/journal.ppat.1010784. eCollection 2022 Sep. PLoS Pathog. 2022. PMID: 36074792 Free PMC article. Review. - Modulation of yeast Erg1 expression and terbinafine susceptibility by iron bioavailability.
Jordá T, Martínez-Martín A, Martínez-Pastor MT, Puig S. Jordá T, et al. Microb Biotechnol. 2022 Nov;15(11):2705-2716. doi: 10.1111/1751-7915.14102. Epub 2022 Jul 15. Microb Biotechnol. 2022. PMID: 35837730 Free PMC article. - Heterologous Expression and Biochemical Characterization of the Human Zinc Transporter 1 (ZnT1) and Its Soluble C-Terminal Domain.
Cotrim CA, Jarrott RJ, Whitten AE, Choudhury HG, Drew D, Martin JL. Cotrim CA, et al. Front Chem. 2021 Apr 30;9:667803. doi: 10.3389/fchem.2021.667803. eCollection 2021. Front Chem. 2021. PMID: 33996761 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources