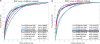A Deep Convolutional Neural Network for segmenting and classifying epithelial and stromal regions in histopathological images - PubMed (original) (raw)
A Deep Convolutional Neural Network for segmenting and classifying epithelial and stromal regions in histopathological images
Jun Xu et al. Neurocomputing (Amst). 2016.
Abstract
Epithelial (EP) and stromal (ST) are two types of tissues in histological images. Automated segmentation or classification of EP and ST tissues is important when developing computerized system for analyzing the tumor microenvironment. In this paper, a Deep Convolutional Neural Networks (DCNN) based feature learning is presented to automatically segment or classify EP and ST regions from digitized tumor tissue microarrays (TMAs). Current approaches are based on handcraft feature representation, such as color, texture, and Local Binary Patterns (LBP) in classifying two regions. Compared to handcrafted feature based approaches, which involve task dependent representation, DCNN is an end-to-end feature extractor that may be directly learned from the raw pixel intensity value of EP and ST tissues in a data driven fashion. These high-level features contribute to the construction of a supervised classifier for discriminating the two types of tissues. In this work we compare DCNN based models with three handcraft feature extraction based approaches on two different datasets which consist of 157 Hematoxylin and Eosin (H&E) stained images of breast cancer and 1376 immunohistological (IHC) stained images of colorectal cancer, respectively. The DCNN based feature learning approach was shown to have a F1 classification score of 85%, 89%, and 100%, accuracy (ACC) of 84%, 88%, and 100%, and Matthews Correlation Coefficient (MCC) of 86%, 77%, and 100% on two H&E stained (NKI and VGH) and IHC stained data, respectively. Our DNN based approach was shown to outperform three handcraft feature extraction based approaches in terms of the classification of EP and ST regions.
Keywords: Breast histopathology; Colorectal cancer; Deep Convolutional Neural Networks; Feature representation; The classification of epithelial and stromal regions.
Figures
Fig. 1
The architecture of the new DCNN employed in this work. The approach comprises of (a) two alternating convolutional layers (or C-layers) with the convolutional operation (b) and max-pooling layers (or P-layers) with the max-pooling operation, and (c) two full connection layers (or FC-layers), and an output layer.
Fig. 2
The illustration of DCNN+SMC approach for Epithelial and Stromal segmentation and classification for H&E (a–f) and IHC (g–k) stained histologic images. The original H&E (a) and IHC (g) stained images are over-segmented into sub-images using a SLIC (b) and fixed-size square window based approach (h), respectively. An exemplar patch (c) is resized into smaller 50 × 50 sub-images (d). The sub-images (d and i) are then fed to a DCNN (e and j) for segmentation and classification of epithelial and stromal regions, shown in panels (f) and (k), respectively. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
Fig. 3
Segmentation of epithelial (red) and stromal (green) regions on a tissue image (a) using the different segmentation approaches on _D_1. (b) The ground truth (a) annotations of stromal and epithelial regions by an expert pathologist. The classification results are shown for DCNN-Ncut-SVM (c), DCNN-Ncut-SMC (d), DCNN-SLIC-SVM (e), DCNN-SLIC-SMC (f), DCNN-SW-SVM (g), DCNN-SW-SMC (h), and Color-SW-SVM (i), respectively. (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
Fig. 4
The probability maps rendered by the different DCNN based approaches (Columns 3 and 4) and [19] (in Column 2) for classifying EP ((a)–(e) in the left block, Column 1) and ST ((f)–(k) in the right block, Column 1) patches on _D_2. The false-color (defined by the heat map (l)) of sub-images in Columns 2–4 reflect the confidence score in predicting them as EP/ST regions via Linda [19], DCNN+SVM, and DCNN+SMC, respectively. The various colors in the heat map (l) correspond to the predicted confidence scores (red=EP with 100% likelihood and blue=ST with 100% likelihood). (For interpretation of the references to color in this figure caption, the reader is referred to the web version of this paper.)
Fig. 5
The ROC curves for the different models (see Table 3) for detecting EP and ST regions on NKI (a) and VGH (b) data cohorts within _D_1.
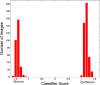
Fig. 6
A plot of the score value on the _X_-axis versus the number of image patches on the _Y_-axis for the model DCNN-SW-SVM on dataset _D_2.
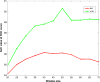
Fig. 7
Plot of AUC versus window size for the DCNN-SW-SVM model on NKI and VGH datasets within _D_1.
Similar articles
- Automated segmentation of cell membranes to evaluate HER2 status in whole slide images using a modified deep learning network.
Khameneh FD, Razavi S, Kamasak M. Khameneh FD, et al. Comput Biol Med. 2019 Jul;110:164-174. doi: 10.1016/j.compbiomed.2019.05.020. Epub 2019 May 30. Comput Biol Med. 2019. PMID: 31163391 - A convolution neural network with multi-level convolutional and attention learning for classification of cancer grades and tissue structures in colon histopathological images.
Dabass M, Vashisth S, Vig R. Dabass M, et al. Comput Biol Med. 2022 Aug;147:105680. doi: 10.1016/j.compbiomed.2022.105680. Epub 2022 Jun 2. Comput Biol Med. 2022. PMID: 35671654 - Deep learning with convolutional neural network in the assessment of breast cancer molecular subtypes based on US images: a multicenter retrospective study.
Jiang M, Zhang D, Tang SC, Luo XM, Chuan ZR, Lv WZ, Jiang F, Ni XJ, Cui XW, Dietrich CF. Jiang M, et al. Eur Radiol. 2021 Jun;31(6):3673-3682. doi: 10.1007/s00330-020-07544-8. Epub 2020 Nov 23. Eur Radiol. 2021. PMID: 33226454 - Psoriasis skin biopsy image segmentation using Deep Convolutional Neural Network.
Pal A, Garain U, Chandra A, Chatterjee R, Senapati S. Pal A, et al. Comput Methods Programs Biomed. 2018 Jun;159:59-69. doi: 10.1016/j.cmpb.2018.01.027. Epub 2018 Feb 6. Comput Methods Programs Biomed. 2018. PMID: 29650319 - Breast cancer cell nuclei classification in histopathology images using deep neural networks.
Feng Y, Zhang L, Yi Z. Feng Y, et al. Int J Comput Assist Radiol Surg. 2018 Feb;13(2):179-191. doi: 10.1007/s11548-017-1663-9. Epub 2017 Aug 31. Int J Comput Assist Radiol Surg. 2018. PMID: 28861708 Review.
Cited by
- Discovery and generalization of tissue structures from spatial omics data.
Wu Z, Kondo A, McGrady M, Baker EAG, Chidester B, Wu E, Rahim MK, Bracey NA, Charu V, Cho RJ, Cheng JB, Afkarian M, Zou J, Mayer AT, Trevino AE. Wu Z, et al. Cell Rep Methods. 2024 Aug 19;4(8):100838. doi: 10.1016/j.crmeth.2024.100838. Epub 2024 Aug 9. Cell Rep Methods. 2024. PMID: 39127044 Free PMC article. - The development and validation of pathological sections based U-Net deep learning segmentation model for the detection of esophageal mucosa and squamous cell neoplasm.
Su F, Zhang W, Liu Y, Chen S, Lin M, Feng M, Yin J, Tan L, Shen Y. Su F, et al. J Gastrointest Oncol. 2023 Oct 31;14(5):1982-1992. doi: 10.21037/jgo-23-587. Epub 2023 Sep 29. J Gastrointest Oncol. 2023. PMID: 37969831 Free PMC article. - Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis.
Litjens G, Sánchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, Hulsbergen-van de Kaa C, Bult P, van Ginneken B, van der Laak J. Litjens G, et al. Sci Rep. 2016 May 23;6:26286. doi: 10.1038/srep26286. Sci Rep. 2016. PMID: 27212078 Free PMC article. - Metabolic light absorption, scattering, and emission (MetaLASE) microscopy.
Restall BS, Haven NJM, Martell MT, Cikaluk BD, Wang J, Kedarisetti P, Tejay S, Adam BA, Sutendra G, Li X, Zemp RJ. Restall BS, et al. Sci Adv. 2024 Oct 18;10(42):eadl5729. doi: 10.1126/sciadv.adl5729. Epub 2024 Oct 18. Sci Adv. 2024. PMID: 39423271 Free PMC article. - Machine Learning Methods for Histopathological Image Analysis.
Komura D, Ishikawa S. Komura D, et al. Comput Struct Biotechnol J. 2018 Feb 9;16:34-42. doi: 10.1016/j.csbj.2018.01.001. eCollection 2018. Comput Struct Biotechnol J. 2018. PMID: 30275936 Free PMC article. Review.
References
- Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Susstrunk S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell. 2012;34(11):2274–2282. - PubMed
- Ali S, et al. Spatially aware cell clusters graphs: predicting outcome in HPV associated oropharyngeal tumors. Medical Image Computing and Computer-Assisted Intervention. 2013;8149:412–519. - PubMed
- Amaral T, McKenna S, Robertson K, Thompson A. Classification and immunohistochemical scoring of breast tissue microarray spots. IEEE Trans Biomed Eng. 2013 Oct;60(10):2806–2814. - PubMed
- Beck AH, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108ra113. - PubMed
- Bengio Y, Lamblin P, Popovici D, Larochelle H, et al. Greedy layer-wise training of deep networks. Adv Neural Inf Process Syst. 2007;19:153.
Grants and funding
- R01 CA202752/CA/NCI NIH HHS/United States
- R21 CA167811/CA/NCI NIH HHS/United States
- R01 CA136535/CA/NCI NIH HHS/United States
- R43 EB015199/EB/NIBIB NIH HHS/United States
- U24 CA199374/CA/NCI NIH HHS/United States
- R21 CA179327/CA/NCI NIH HHS/United States
- R01 CA140772/CA/NCI NIH HHS/United States
- R21 CA195152/CA/NCI NIH HHS/United States
- R01 DK098503/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous