Colonic Microbiota Encroachment Correlates With Dysglycemia in Humans - PubMed (original) (raw)
Colonic Microbiota Encroachment Correlates With Dysglycemia in Humans
Benoit Chassaing et al. Cell Mol Gastroenterol Hepatol. 2017.
Abstract
Background and aims: Mucoid structures that coat the epithelium play an essential role in keeping the intestinal microbiota at a safe distance from host cells. Encroachment of bacteria into the normally almost-sterile inner mucus layer has been observed in inflammatory bowel disease and in mouse models of colitis. Moreover, such microbiota encroachment has also been observed in mouse models of metabolic syndrome, which are associated low-grade intestinal inflammation. Hence, we investigated if microbiota encroachment might correlate with indices of metabolic syndrome in humans.
Methods: Confocal microscopy was used to measure bacterial-epithelial distance of the closest bacteria per high-powered field in colonic biopsies of all willing participants undergoing cancer screening colonoscopies.
Results: We observed that, among all subjects, bacterial-epithelial distance was inversely correlated with body mass index, fasting glucose levels, and hemoglobin A1C. However, this correlation was driven by dysglycemic subjects, irrespective of body mass index, whereas the difference in bacterial-epithelial distance between obese and nonobese subjects was eliminated by removal of dysglycemic subjects.
Conclusions: We conclude that microbiota encroachment is a feature of insulin resistance-associated dysglycemia in humans.
Keywords: BMI, body mass index; HPF, high-powered field; IBD, inflammatory bowel disease; Metabolic Syndrome; Microbiota; Mucus Layer; PBS, phosphate-buffered saline; TLR, Toll-like receptor.
Figures
Graphical abstract
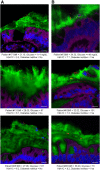
Figure 1
Microbiota localization in human with or without metabolic syndrome. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution. Representative images of confocal microscopy analysis of microbiota localization; Muc2 (green), actin (purple), bacteria (red), and DNA (blue). (A) Patients without diabetes mellitus. (B) Patients with diabetes mellitus. Bar = 10 μm; n = 42. HbA1C, hemoglobin A1C.
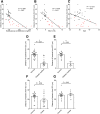
Figure 2
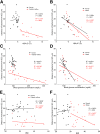
Metabolic syndrome correlates with microbiota encroachment in human. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution, followed by confocal microscopy analysis of microbiota localization. (A) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus fasting blood glucose concentration. (B) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus hemoglobin A1C level. (C) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus body mass index. (D–G) Distances of the closest bacteria to intestinal epithelial cells per condition over 5 high-powered field according to the diabetes mellitus (D, E) or the obese (F, G) status. In E, obese patients were removed from the analysis. In G, patients with diabetes mellitus were removed from the analysis. Linear regression line was plotted and _R_2 and P values were determined. Significance was determined by Student t test. *P < .05); n = 42; red dots represent subjects with diabetes. HbA1C, hemoglobin A1C; IEC, intestinal epithelial cells.
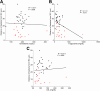
Figure 3
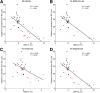
Circulating lipid profile does not correlate with microbiota encroachment in human. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution, followed by confocal microscopy analysis of microbiota localization. (A) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus cholesterol concentration. Linear regression line was plotted and _R_2 and P values were determined. (B) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus triglyceride concentration. Linear regression line was plotted and _R_2 and P values were determined. (C) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus low-density lipoprotein concentration. Linear regression line was plotted and _R_2 and P values were determined. n = 42; red dots represent subjects with diabetes. IEC, intestinal epithelial cells; LDL, low-density lipoprotein.
Figure 4
Dysglycemia correlates with microbiota encroachment in human with diabetes. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution, followed by confocal microscopy analysis of microbiota localization. Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus hemoglobin A1C level (A, B), fasting blood glucose concentration (C, D), or body mass index (E, F). Linear regression line was plotted and _R_2 and P values were determined. n = 42; red dots represent subjects with diabetes (A, C, and E) or obese subjects (B, D, and F). HbA1C, hemoglobin A1C; IEC, intestinal epithelial cells.
Figure 5
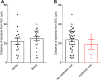
Ethnicity or antibiotic use do not correlate with microbiota encroachment in human. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution, followed by confocal microscopy analysis of microbiota localization. (A) Distances of the closest bacteria to intestinal epithelial cells per condition over 5 high-powered fields according to the ethnicity. (B) Distances of the closest bacteria to intestinal epithelial cells per condition over 5 high-powered fields according to the antibiotic use status. IEC, intestinal epithelial cells.
Figure 6
Antidiabetic drug use does not impact microbiota encroachment in human. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution, followed by confocal microscopy analysis of microbiota localization. (A) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus hemoglobin A1C level. Linear regression line was plotted and _R_2 and P values were determined. (B) Patients using metformin drug were removed from the analysis. (C) Patients using insulin drug were removed from the analysis. (D) Patients using glipizide drug were removed from the analysis. Linear regression line was plotted and _R_2 and P values were determined. n = 42; red dots represent subjects with diabetes. HbA1C, hemoglobin A1C; IEC, intestinal epithelial cells.
Figure 7
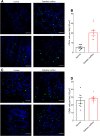
Metabolic syndrome correlates with increased CD19 + cell population in the intestinal mucosa. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution. Representative images of confocal microscopy analysis of CD19 (A) and CD68 (C) staining (green) and DNA (blue). Number of CD19+ (B) and CD68+ (D) cells per field (0.102 mm2). Significance was determined by Student t test. *P < .05). Bar = 50 μm; n = 7.
Figure 8
Streptozotocin-induced type 1 diabetes and associated increased fecal glucose is not sufficient to induce microbiota encroachment in mice. Type 1 diabetes was induced in mice by streptozotocin injection for 5 consecutive days. (A) Body weight over time. (B) Five-hour fasting blood glucose concentration on Day 0 and Day 15. (C) Day 0 and Day 15 fecal glucose concentration. (D) Representative images of confocal microscopy analysis of microbiota localization; Muc2 (green), actin (purple), bacteria (red), and DNA (blue). (E) Distances of the closest bacteria to intestinal epithelial cells per condition over 5 high-powered fields according to the diabetes mellitus status. Significance was determined by Student t test (*P < .05). Bar = 50 μm; n = 5–9. IEC, intestinal epithelial cells; STZ, streptozotocin.
Similar articles
- Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome.
Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, Gewirtz AT. Chassaing B, et al. Nature. 2015 Mar 5;519(7541):92-6. doi: 10.1038/nature14232. Epub 2015 Feb 25. Nature. 2015. PMID: 25731162 Free PMC article. - Enteric Delivery of Regenerating Family Member 3 alpha Alters the Intestinal Microbiota and Controls Inflammation in Mice With Colitis.
Darnaud M, Dos Santos A, Gonzalez P, Augui S, Lacoste C, Desterke C, De Hertogh G, Valentino E, Braun E, Zheng J, Boisgard R, Neut C, Dubuquoy L, Chiappini F, Samuel D, Lepage P, Guerrieri F, Doré J, Bréchot C, Moniaux N, Faivre J. Darnaud M, et al. Gastroenterology. 2018 Mar;154(4):1009-1023.e14. doi: 10.1053/j.gastro.2017.11.003. Epub 2017 Nov 11. Gastroenterology. 2018. PMID: 29133078 - Dismicrobism in inflammatory bowel disease and colorectal cancer: changes in response of colocytes.
Tomasello G, Tralongo P, Damiani P, Sinagra E, Di Trapani B, Zeenny MN, Hussein IH, Jurjus A, Leone A. Tomasello G, et al. World J Gastroenterol. 2014 Dec 28;20(48):18121-30. doi: 10.3748/wjg.v20.i48.18121. World J Gastroenterol. 2014. PMID: 25561781 Free PMC article. Review. - Bacterial immune interaction in experimental colitis.
Xue LY, Ouyang Q, Zhou XG, Huang ZH, Chen W, Chen M, Yu LM. Xue LY, et al. J Dig Dis. 2013 Oct;14(10):526-35. doi: 10.1111/1751-2980.12079. J Dig Dis. 2013. PMID: 23734583 - The Intestinal Microbiota in Inflammatory Bowel Disease.
Becker C, Neurath MF, Wirtz S. Becker C, et al. ILAR J. 2015;56(2):192-204. doi: 10.1093/ilar/ilv030. ILAR J. 2015. PMID: 26323629 Review.
Cited by
- A Saudi Heart Association Position Statement on Obesity and Cardiovascular Disease.
Alhabeeb W, Kinsara AJ, Bakhsh A, Tash A, Alshammary A, Almasood A, Alghalayini K, Arafah M, Hamdy O, Alsifri S, Kharabsheh SM, Alkattan W. Alhabeeb W, et al. J Saudi Heart Assoc. 2024 Oct 2;36(3):263-300. doi: 10.37616/2212-5043.1391. eCollection 2024. J Saudi Heart Assoc. 2024. PMID: 39469000 Free PMC article. - Dietary and metabolic effects on intestinal stem cells in health and disease.
Shay JES, Yilmaz ÖH. Shay JES, et al. Nat Rev Gastroenterol Hepatol. 2024 Oct 2. doi: 10.1038/s41575-024-00980-7. Online ahead of print. Nat Rev Gastroenterol Hepatol. 2024. PMID: 39358589 Review. - A history of repeated antibiotic usage leads to microbiota-dependent mucus defects.
Krigul KL, Feeney RH, Wongkuna S, Aasmets O, Holmberg SM, Andreson R, Puértolas-Balint F, Pantiukh K, Sootak L, Org T, Tenson T, Org E, Schroeder BO. Krigul KL, et al. Gut Microbes. 2024 Jan-Dec;16(1):2377570. doi: 10.1080/19490976.2024.2377570. Epub 2024 Jul 21. Gut Microbes. 2024. PMID: 39034613 Free PMC article. - Heavy arch: from inflammatory bowel diseases to metabolic disorders.
Adolph TE, Meyer M, Jukic A, Tilg H. Adolph TE, et al. Gut. 2024 Jul 11;73(8):1376-1387. doi: 10.1136/gutjnl-2024-331914. Gut. 2024. PMID: 38777571 Free PMC article. Review. - Human milk oligosaccharide 2'-fucosyllactose protects against high-fat diet-induced obesity by changing intestinal mucus production, composition and degradation linked to changes in gut microbiota and faecal proteome profiles in mice.
Paone P, Latousakis D, Terrasi R, Vertommen D, Jian C, Borlandelli V, Suriano F, Johansson MEV, Puel A, Bouzin C, Delzenne NM, Salonen A, Juge N, Florea BI, Muccioli GG, Overkleeft H, Van Hul M, Cani PD. Paone P, et al. Gut. 2024 Sep 9;73(10):1632-1649. doi: 10.1136/gutjnl-2023-330301. Gut. 2024. PMID: 38740509 Free PMC article.
References
- Hooper L.V., Macpherson A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. - PubMed
- Chassaing B., Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. - PubMed
- Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. - PubMed
Grants and funding
- R01 DK061417/DK/NIDDK NIH HHS/United States
- R01 DK080684/DK/NIDDK NIH HHS/United States
- R01 DK083890/DK/NIDDK NIH HHS/United States
- R01 DK099071/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources