Targeting autophagy in cancer - PubMed (original) (raw)
Review
. 2017 Sep;17(9):528-542.
doi: 10.1038/nrc.2017.53. Epub 2017 Jul 28.
Affiliations
- PMID: 28751651
- PMCID: PMC5975367
- DOI: 10.1038/nrc.2017.53
Review
Targeting autophagy in cancer
Jean M Mulcahy Levy et al. Nat Rev Cancer. 2017 Sep.
Abstract
Autophagy is a mechanism by which cellular material is delivered to lysosomes for degradation, leading to the basal turnover of cell components and providing energy and macromolecular precursors. Autophagy has opposing, context-dependent roles in cancer, and interventions to both stimulate and inhibit autophagy have been proposed as cancer therapies. This has led to the therapeutic targeting of autophagy in cancer to be sometimes viewed as controversial. In this Review, we suggest a way forwards for the effective targeting of autophagy by understanding the context-dependent roles of autophagy and by capitalizing on modern approaches to clinical trial design.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing interests.
Figures
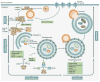
Figure 1. Timeline of the Major Discoveries Leading to the Successful Targeting of Autophagy in Cancer
De Duve first coined the term ‘autophagy’ during a lysosomal conference in 1963. Since then key discoveries have been made elucidating the mechanisms of the process from yeast to cultured cell lines, into mice, and finally culminating in successful clinical trials and case studies in patient tumors. The timeline concludes with the Nobel Prize awarded to Yoshinori Ohsumi for Physiology or Medicine in 2016, emphasizing the impact of his work as well as that of many others along the way. CQ, chloroquine; HCQ, hydroxychloroquine 1963 - Christian de Duve coins the term “autophagy” 1970 - Bedoya shows that the anti-malarial drug, CQ can inhibit tumour cell growth in vitro 1997 - The Ohsumi group clone the first autophagy specific gene, ATG1, from Saccharomyces cerevisiae, 1998 - Mizushima and colleagues identify the first autophagy specific gene in higher eukaryotes 1998 - Murakami and colleagues observe that CQ can inhibit autophagy 1999 - The Levine group suggest that Beclin 1 is a tumor suppressor gene 2003 - Briceno et al. show that CQ has anti-tumour affects in patients with glioblastoma 2004 - The Mizushima group create the first autophagy deficient mouse (_Atg5_−/−), implicating autophagy in development 2007 - Amaravadi and colleagues show that inhibition of autophagy has a combinatory effect with other anti-cancer drugs 2013 - A number of groups show that genetic knock out of autophagy-related genes in tumour cells decreases tumour growth in vivo, e.g. ,, 2014 - The White group show that systemic genetic inhibition of autophagy in tumour-bearing mice can create a therapeutic window to treat lung cancer 2014 - Multiple groups publish data from Phase I/II clinical trials using HCQ to selectively target autophagy in cancer patients– 2016 - Yoshinori Ohsumi is awarded the Nobel Prize for Physiology or Medicine for his work discovering the mechanisms of autophagy 2017 - Autophagy inhibition can overcome resistance to kinase inhibitors in tumour cells and in patients
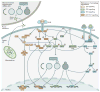
Figure 2. Autophagy can be inhibited at multiple stages
The process of autophagy is divided into five distinct stages: initiation, vesicle nucleation, vesicle elongation, vesicle fusion and cargo degradation. Nonspecific macroautophagy is initiated by upstream activation through either nutrient starvation or growth factors. Under starvation conditions, a drop in glucose transport results in a release of mTOR inhibition of the ULK1 complex, allowing for the progression of autophagy. The ULK1 complex (comprising ULK1, ULK2, FIP200, ATG101 and ATG13) induces vesicle nucleation which is then mediated by a class III PI3K complex consisting of multiple proteins. Beclin 1, a BCL-2 homology (BH)-3 domain only protein, is phosphorylated by ULK1 and acts as an overall scaffold for the PI3K complex, facilitating localization of autophagic proteins to the phagophore. BCL-2 and BCL-XL interact with Beclin 1 at the BH3 domain to decrease the pro-autophagic activity of Beclin 1 by interrupting the Beclin 1–VPS34 complex formation and decreasing the interaction of Beclin 1 with UVRAG. Additional negative regulation of this process occurs with the phosphorylation of VPS34 (also known as PIK3C3), which decreases its interaction with Beclin 1. In contrast, AMBRA binds Beclin 1 and stabilizes the PI3K complex. ATG14 and UVRAG also bind Beclin 1 to promote interactions between Beclin 1 and VPS34 and phagophore formation. VPS15 is required for optimal VPS34 function by enhancing VPS34 interaction with Beclin 1. The growing double membrane undergoes vesicle elongation to eventually form an autophagasome: a process mediated by two ubiquitin-like conjugation systems. The first system involves the conjugation of phosphatidylethanolamine (PE) to cytoplasmic LC3-I to generate the lipidated form, LC3-II which is facilitated by the protease, ATG4B, and the E1-like enzyme, ATG7, whereby LC3-II is incorporated into the growing membrane. The second conjugation system is mediated again by ATG7 as well as the E2-like enzyme, ATG10, resulting in an ATG5-ATG12 conjugate. Subsequently, the SNARE protein, syntaxin 17 (STX17) facilitates autophagosome fusion with the lysosome, resulting in an autophagolysosome. The low pH of the lysosome results in degradation of the autophagosome contents. This process can be targeted pharmacologically upstream by means of direct ULK1, VPS34, or Beclin 1 inhibition. It can also be targeted by wortmannin and 3-methyladenine (3-MA) which act as PI3K inhibitors. Downstream targets include direct ATG4B inhibitors as well as chloroquine or hydroxychloroquine and bafilomycin, which act to prevent autophagosome fusion with the lysosome. PE, phosphatidylethanolamine.
Figure 3. Molecular Mechanism of Autophagy Dependence
Pre-clinical and clinical models have indicated that the tumor microenvironment, for example pancreatic stellate cells in the case of pancreatic cancer, p53 status, RAS family status, activation of JAK–STAT and PI3K signaling may all play roles in the determination of autophagy dependence within cancer cells, both in vitro and in patients. These pathways have all been shown to affect autophagy either positively or negatively and many participate in cross-pathway signaling. Signaling through p53 can both promote and inhibit autophagy and may interact with other proteins activated by mutations to enhance autophagy dependence, especially in pancreatic cancer. Activation of EGFR via amplification or mutation leads to the downstream up-regulation of the PI3K-AKT-mTOR pathway as well as activation of STAT3 and the RAS pathway. Although autophagy inhibition can occur through mTOR activation, these downstream effects collectively result in stimulation of autophagy and an increase in autophagy dependence. Mutations or alterations in the RAS family (specifically KRAS) have been shown to promote autophagy, enhancing tumor growth and therapy resistance. Specific mutations in RAF such as _BRAF_V600E promote autophagy dependence in multiple tumors including central nervous system (CNS) tumors and melanoma. Finally, autophagy regulation of JAK-STAT signaling through IL-6 has been identified as a mechanism of autophagy dependence in breast cancer. All of these pathways are complex and interact on multiple levels. Identification of tumors with these pathways and as of yet to be identified pathways will provide methods of detection of autophagy dependent tumors.
Similar articles
- Targeting Autophagy in Cancer: Update on Clinical Trials and Novel Inhibitors.
Chude CI, Amaravadi RK. Chude CI, et al. Int J Mol Sci. 2017 Jun 16;18(6):1279. doi: 10.3390/ijms18061279. Int J Mol Sci. 2017. PMID: 28621712 Free PMC article. Review. - Autophagy therapeutics: preclinical basis and initial clinical studies.
Zhan L, Li J, Wei B. Zhan L, et al. Cancer Chemother Pharmacol. 2018 Dec;82(6):923-934. doi: 10.1007/s00280-018-3688-3. Epub 2018 Sep 17. Cancer Chemother Pharmacol. 2018. PMID: 30225602 Review. - Chloroquine and hydroxychloroquine in antitumor therapies based on autophagy-related mechanisms.
Ferreira PMP, Sousa RWR, Ferreira JRO, Militão GCG, Bezerra DP. Ferreira PMP, et al. Pharmacol Res. 2021 Jun;168:105582. doi: 10.1016/j.phrs.2021.105582. Epub 2021 Mar 26. Pharmacol Res. 2021. PMID: 33775862 Review. - Therapeutic Targeting of Autophagy.
Towers CG, Thorburn A. Towers CG, et al. EBioMedicine. 2016 Dec;14:15-23. doi: 10.1016/j.ebiom.2016.10.034. Epub 2016 Oct 23. EBioMedicine. 2016. PMID: 28029600 Free PMC article. Review. - The role of autophagy in cancer: therapeutic implications.
Yang ZJ, Chee CE, Huang S, Sinicrope FA. Yang ZJ, et al. Mol Cancer Ther. 2011 Sep;10(9):1533-41. doi: 10.1158/1535-7163.MCT-11-0047. Epub 2011 Aug 30. Mol Cancer Ther. 2011. PMID: 21878654 Free PMC article. Review.
Cited by
- T1/T2 Proportional Magnetic Resonance Nanoprobe Monitoring Tumor Autophagy during Chemotherapy.
Cui J, Zhang T, Wang F, Feng L, Deng G, Wu T, Yin L, Hu Y. Cui J, et al. Nanomaterials (Basel). 2024 Oct 18;14(20):1673. doi: 10.3390/nano14201673. Nanomaterials (Basel). 2024. PMID: 39453009 Free PMC article. - Altered Protein and Gene Expression of Beclin-1 Correlates with Poor Prognosis of Hcv-Associated Hepatocellular Carcinoma in Egyptian Patients.
Ehsan NA, Mosbeh AM, Elkhadry SW, Gomaa AI, Elsabaawy MM, Elazab DS. Ehsan NA, et al. Asian Pac J Cancer Prev. 2021 Apr 1;22(4):1115-1122. doi: 10.31557/APJCP.2021.22.4.1115. Asian Pac J Cancer Prev. 2021. PMID: 33906303 Free PMC article. - Role of the mesenchymal stem cells derived from adipose tissue in changing the rate of breast cancer cell proliferation and autophagy, in vitro and in vivo.
Adelipour M, Allameh A, Sheikhi A, Ranjbaran M, Naghashpour M, Nazeri Z, Mojiri-Forushani H, Golabi S. Adelipour M, et al. Iran J Basic Med Sci. 2021 Jan;24(1):98-107. doi: 10.22038/ijbms.2020.51461.11678. Iran J Basic Med Sci. 2021. PMID: 33643577 Free PMC article. - The endoplasmic reticulum membrane protein Sec62 as potential therapeutic target in SEC62 overexpressing tumors.
Zimmermann JSM, Linxweiler J, Radosa JC, Linxweiler M, Zimmermann R. Zimmermann JSM, et al. Front Physiol. 2022 Oct 3;13:1014271. doi: 10.3389/fphys.2022.1014271. eCollection 2022. Front Physiol. 2022. PMID: 36262254 Free PMC article. Review. - Cuproptosis-related gene index: A predictor for pancreatic cancer prognosis, immunotherapy efficacy, and chemosensitivity.
Huang X, Zhou S, Tóth J, Hajdu A. Huang X, et al. Front Immunol. 2022 Aug 25;13:978865. doi: 10.3389/fimmu.2022.978865. eCollection 2022. Front Immunol. 2022. PMID: 36090999 Free PMC article.
References
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. - PubMed
- The Nobel Assembly. The Nobel Assembly at Karolinska Instiutet has today decided to award the 2016 Nobel Prize in Physiology or Medicine to Yoshinori Ohsumi. 2016 < https://www.nobelprize.org/nobel_prizes/medicine/laureates/2016/press.html>.
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA190216/CA/NCI NIH HHS/United States
- R01 CA150925/CA/NCI NIH HHS/United States
- K08 CA193982/CA/NCI NIH HHS/United States
- R21 CA197887/CA/NCI NIH HHS/United States
- R01 CA190170/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources