Age-associated dysregulation of protein metabolism in the mammalian oocyte - PubMed (original) (raw)
. 2017 Dec;16(6):1381-1393.
doi: 10.1111/acel.12676. Epub 2017 Oct 10.
Affiliations
- PMID: 28994181
- PMCID: PMC5676066
- DOI: 10.1111/acel.12676
Age-associated dysregulation of protein metabolism in the mammalian oocyte
Francesca E Duncan et al. Aging Cell. 2017 Dec.
Abstract
Reproductive aging is characterized by a marked decline in oocyte quality that contributes to infertility, miscarriages, and birth defects. This decline is multifactorial, and the underlying mechanisms are under active investigation. Here, we performed RNA-Seq on individual growing follicles from reproductively young and old mice to identify age-dependent functions in oocytes. This unbiased approach revealed genes involved in cellular processes known to change with age, including mitochondrial function and meiotic chromosome segregation, but also uncovered previously unappreciated categories of genes related to proteostasis and organelles required for protein metabolism. We further validated our RNA-Seq data by comparing nucleolar structure and function in oocytes from reproductively young and old mice, as this organelle is central for protein production. We examined key nucleolar markers, including upstream binding transcription factor (UBTF), an RNA polymerase I cofactor, and fibrillarin, an rRNA methyltransferase. In oocytes from mice of advanced reproductive age, UBTF was primarily expressed in giant fibrillar centers (GFCs), structures associated with high levels of rDNA transcription, and fibrillarin expression was increased ~2-fold. At the ultrastructural level, oocyte nucleoli from reproductively old mice had correspondingly more prominent fibrillar centers and dense fibrillar centers relative to young controls and more ribosomes were found in the cytoplasm. Taken together, our findings are significant because the growing oocyte is one of the most translationally active cells in the body and must accumulate high-quality maternally derived proteins to support subsequent embryo development. Thus, perturbations in protein metabolism are likely to have a profound impact on gamete health.
Keywords: folliculogenesis; nucleolus; oogenesis; proteostasis; reproductive aging; ribosome.
© 2017 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures
Figure 1
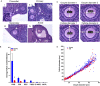
Reproductive aging in CB6F1 mice is associated with reduced ovarian reserve and altered follicle growth dynamics. (A) Representative hematoxylin and eosin (H&E)‐stained ovarian tissue sections demonstrate follicle‐stage classes according to histologic morphology (see Fig. S1 for additional images). White arrows highlight the particular follicle class. Insets show magnified images of primordial and primary follicles. Scale bars are (I–III) 100 μm and (IV) 200 μm. (B) Graph showing the average number of each follicle class per ovarian section (every fifth section of serially sectioned ovaries was counted; N = 3 ovaries from reproductively young and old mice). PRD: primordial; PRI: primary; SEC: secondary; T‐SEC: transitioning secondary; E‐ANTL: early antral; ANTL: antral. Two‐way
anova
was performed; asterisks denote P < 0.001. (C) Images showing how oocyte and follicle diameters were determined by the mean of two perpendicular measurements on H&E‐stained ovarian tissue sections at 40× magnification. (D) Mean oocyte and follicle diameters were plotted for each individual growing follicle (primary–transitioning secondary). Each dot shows the mean oocyte diameter on the _x_‐axis, and its corresponding follicle diameter on the _y_‐axis. For each ovary, a minimum of 50 follicles in each stage were measured. Pearson's correlation and linear regression were performed. R 2 for young and old were 0.9455 and 0.9256, respectively. The slopes between age cohorts were significantly different (P = 0.039).
Figure 2
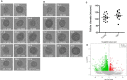
RNA‐Seq on individual follicles from reproductively young and old mice reveals differential gene expression despite indistinguishable morphology. Brightfield microscopy images of all follicles from reproductively (A) young and (B) old mice, which were processed for individual follicle RNA sequencing and analysis. Each row shows follicles that were isolated from an individual mouse. Each follicle's unique identifier used throughout the manuscript is listed at the top left corner of each image. The mean diameter (μm) for each follicle, determined by taking the mean of two perpendicular measurements, is shown at the bottom of each image. The scale bar is 50 μm. (C) The graph shows the mean follicle diameter measurement of each follicle shown in (A) and (B). A _t_‐test was performed and there was no difference in diameters between the age cohorts (P = 0.2338). (D) Genes that are overall differentially expressed between follicles from reproductively young and old mice are shown in a volcano plot. Genes that are expressed more highly in young are shown in red (411), and those that are more highly expressed in old are shown in green (1553).
Figure 3
Principle component analysis can distinguish follicles from reproductively young and old mice and follicles from reproductively old mice exhibit greater variability in gene expression. (A) The first three components of PCA effectively separate the young samples (red, orange, yellow, green) from the old (blues and purples). Two follicles from reproductively old mice are closer to the young than the other old (O16.3, O17.5). (B) PCA components 4 and 5 separate follicles derived from each individual old mouse from each other. (C) PCA components 5 and 6 separate follicles derived from each individual young mouse from each other. (D) The pairwise comparisons of the Pearson's correlations, shown collectively in a box plot from young–young (YY), young–old (YO), and old–old (OO) samples, indicate that young samples are more highly correlated with each other than old samples. (E) For young and old samples, we compared CVs of normalized counts for genes detected in all samples. 71% of genes had higher values for old samples, indicating greater variability in gene expression with age. (F) We calculated the number of hypervariable genes per sample. The term ‘hypervariable’ indicates a gene with an extreme expression profile; that is, it has one rpkm _z_‐score > 4. For each sample, we asked how many hypervariable genes had highest expression. Young samples had < 100 on average, while old samples had close to 150 on average, indicating that age was associated with extreme expression.
Figure 4
Genes involved in proteostasis are differentially expressed with age in the growing oocyte. (A) The butterfly plot shows the major trends in significantly enriched GO terms from the differentially expressed genes within the intact follicle. Terms enriched in young samples are on the right, and terms enriched in the old samples are on the left. The _x_‐axis is the –log10 of BH‐corrected _P_‐value. Numbers in bars indicate the total number of differentially expressed genes with the specified term. (B) Genes were assigned to ‘oocyte’ based on their expression in publicly available data. (C) Cellular component GO terms from the differentially expressed genes assigned to the oocyte reveal previously known age‐based signatures (i.e., mitochondria, spindle, microtubules, chromosomes) as well as a new category involved in proteostasis (red).
Figure 5
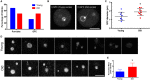
Reproductive aging is associated with altered nucleolar architecture. (A) The patterns of UBTF expression in oocyte nucleoli were examined in follicles isolated from reproductively young and old mice. The graph shows the distribution of UBTF conformations. Chi‐square analysis was performed and P = 0.0097. (B) Representative images of GFC size differences (arrows) in oocyte nucleoli observed in young and old cohorts. The scale bar is 10 μm. (C) The mean GFC diameter in each nucleolus was analyzed by taking the mean of two perpendicular measurements, and these measurements are plotted for the two age cohorts. A _t_‐test was performed, and the asterisks denotes P = 0.0317. Four experimental replicates were performed, and a total of > 15 follicles per group were analyzed. (D). The patterns of fibrillarin expression in oocyte nucleoli were examined in follicles isolated from reproductively young and old mice. Representative images of fibrillarin staining in the nucleoli of oocytes from young (top panel) and old (bottom) cohorts are shown from one experiment. Four experimental replicates were performed. White asterisks indicate the nucleoli that were excluded from quantification to avoid duplication. The scale bar is 10 μm. (E) Fibrillarin intensities were quantified using
image j
, and only a defined central region of each nucleolus was used for quantification. Mean pixel intensity from four independent replicates was normalized to the mean of the control, and the combined fold expression was determined. A _t_‐test was performed, and the asterisks denotes P < 0.0001.
Figure 6
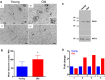
Oocytes from reproductively old mice have increased ribosome numbers. (A) Representative transmission electron microscopy (TEM) images of the cytoplasm of oocytes from reproductively young (I, II) and old (III, IV) mice are shown. Arrows and the boxed region and inset highlight ribosomes. The scale bar is 500 nm. (B) Ribosome number was quantified using
image j
, and the graph shows the average number of ribosomes per defined region of interest in oocytes from reproductively young and old mice. A _t_‐test was performed, and the asterisks denotes P = 0.0139. (C) Oocyte protein extracts from reproductively young and old cohorts were processed for immunoblot analysis with an RPS2 antibody (10 oocytes/lane). The immunoblot was reprobed with an antibody against the oocyte‐specific protein MSY‐2 as a loading control. (D) The graph shows the fold change in RPS2 expression in young and old cohorts from four independent experiments. The overall fold increase in RPS2 expression across experiments was 1.851 (P = 0.0627).
Comment in
- Mammalian oogenesis and female reproductive aging.
Duncan FE, Gerton JL. Duncan FE, et al. Aging (Albany NY). 2018 Feb 5;10(2):162-163. doi: 10.18632/aging.101381. Aging (Albany NY). 2018. PMID: 29410392 Free PMC article. No abstract available.
Similar articles
- Nucleolus-like bodies of fully-grown mouse oocytes contain key nucleolar proteins but are impoverished for rRNA.
Shishova KV, Lavrentyeva EA, Dobrucki JW, Zatsepina OV. Shishova KV, et al. Dev Biol. 2015 Jan 15;397(2):267-81. doi: 10.1016/j.ydbio.2014.11.022. Epub 2014 Dec 4. Dev Biol. 2015. PMID: 25481757 - Immunolocalization of upstream binding factor and pocket protein p130 during final stages of bovine oocyte growth.
Baran V, Pavlok A, Bjerregaard B, Wrenzycki C, Hermann D, Philimonenko VV, Lapathitis G, Hozak P, Niemann H, Motlik J. Baran V, et al. Biol Reprod. 2004 Apr;70(4):877-86. doi: 10.1095/biolreprod.103.018408. Epub 2003 Nov 12. Biol Reprod. 2004. PMID: 14613906 - Ribosomal RNA and nucleolar proteins from the oocyte are to some degree used for embryonic nucleolar formation in cattle and pig.
Maddox-Hyttel P, Svarcova O, Laurincik J. Maddox-Hyttel P, et al. Theriogenology. 2007 Sep 1;68 Suppl 1:S63-70. doi: 10.1016/j.theriogenology.2007.03.015. Epub 2007 Apr 26. Theriogenology. 2007. PMID: 17466364 Review. - The nucleolar protein NOP2 is required for nucleolar maturation and ribosome biogenesis during preimplantation development in mammals.
Wang H, Wang L, Wang Z, Dang Y, Shi Y, Zhao P, Zhang K. Wang H, et al. FASEB J. 2020 Feb;34(2):2715-2729. doi: 10.1096/fj.201902623R. Epub 2019 Dec 24. FASEB J. 2020. PMID: 31908012
Cited by
- Single housing of juveniles accelerates early-stage growth but extends adult lifespan in African turquoise killifish.
Takahashi C, Okabe E, Nono M, Kishimoto S, Matsui H, Ishitani T, Yamamoto T, Uno M, Nishida E. Takahashi C, et al. Aging (Albany NY). 2024 Sep 16;16(18):12443-12472. doi: 10.18632/aging.206111. Epub 2024 Sep 16. Aging (Albany NY). 2024. PMID: 39302208 Free PMC article. - Nucleolar Organizer Regions as Transcription-Based Scaffolds of Nucleolar Structure and Function.
Cockrell AJ, Gerton JL. Cockrell AJ, et al. Results Probl Cell Differ. 2022;70:551-580. doi: 10.1007/978-3-031-06573-6_19. Results Probl Cell Differ. 2022. PMID: 36348121 - Upregulated Ribosomal Pathway Impairs Follicle Development in a Polycystic Ovary Syndrome Mouse Model: Differential Gene Expression Analysis of Oocytes.
Nakanishi N, Osuka S, Kono T, Kobayashi H, Ikeda S, Bayasula B, Sonehara R, Murakami M, Yoshita S, Miyake N, Muraoka A, Kasahara Y, Murase T, Nakamura T, Goto M, Iwase A, Kajiyama H. Nakanishi N, et al. Reprod Sci. 2023 Apr;30(4):1306-1315. doi: 10.1007/s43032-022-01095-7. Epub 2022 Oct 4. Reprod Sci. 2023. PMID: 36194357 - Hyperbaric Oxygen Treatment Ameliorates the Decline in Oocyte Quality and Improves the Fertility of Aged Female Mice.
Ma Y, Zhong Y, Chen X, Liu H, Shi Y, Zhang X, Sun H. Ma Y, et al. Reprod Sci. 2023 Jun;30(6):1834-1840. doi: 10.1007/s43032-022-01082-y. Epub 2022 Dec 15. Reprod Sci. 2023. PMID: 36520404 Free PMC article. - Ribosomal DNA and the nucleolus in the context of genome organization.
Potapova TA, Gerton JL. Potapova TA, et al. Chromosome Res. 2019 Mar;27(1-2):109-127. doi: 10.1007/s10577-018-9600-5. Epub 2019 Jan 17. Chromosome Res. 2019. PMID: 30656516 Review.
References
- Ben‐Meir A, Burstein E, Borrego‐Alvarez A, Chong J, Wong E, Yavorska T, Naranian T, Chi M, Wang Y, Bentov Y, Alexis J, Meriano J, Sung HK, Gasser DL, Moley KH, Hekimi S, Casper RF, Jurisicova A (2015) Coenzyme Q10 restores oocyte mitochondrial function and fertility during reproductive aging. Aging Cell 14, 887–895. - PMC - PubMed
- Bristol‐Gould SK, Kreeger PK, Selkirk CG, Kilen SM, Cook RW, Kipp JL, Shea LD, Mayo KE, Woodruff TK (2006) Postnatal regulation of germ cells by activin: the establishment of the initial follicle pool. Dev. Biol. 298, 132–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 GM104936/GM/NIGMS NIH HHS/United States
- P30 HD002528/HD/NICHD NIH HHS/United States
- S10 RR027564/RR/NCRR NIH HHS/United States
- P20 GM103418/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources