Emerging roles of linker histones in regulating chromatin structure and function - PubMed (original) (raw)
Review
. 2018 Mar;19(3):192-206.
doi: 10.1038/nrm.2017.94. Epub 2017 Oct 11.
Affiliations
- PMID: 29018282
- PMCID: PMC5897046
- DOI: 10.1038/nrm.2017.94
Review
Emerging roles of linker histones in regulating chromatin structure and function
Dmitry V Fyodorov et al. Nat Rev Mol Cell Biol. 2018 Mar.
Abstract
Together with core histones, which make up the nucleosome, the linker histone (H1) is one of the five main histone protein families present in chromatin in eukaryotic cells. H1 binds to the nucleosome to form the next structural unit of metazoan chromatin, the chromatosome, which may help chromatin to fold into higher-order structures. Despite their important roles in regulating the structure and function of chromatin, linker histones have not been studied as extensively as core histones. Nevertheless, substantial progress has been made recently. The first near-atomic resolution crystal structure of a chromatosome core particle and an 11 Å resolution cryo-electron microscopy-derived structure of the 30 nm nucleosome array have been determined, revealing unprecedented details about how linker histones interact with the nucleosome and organize higher-order chromatin structures. Moreover, several new functions of linker histones have been discovered, including their roles in epigenetic regulation and the regulation of DNA replication, DNA repair and genome stability. Studies of the molecular mechanisms of H1 action in these processes suggest a new paradigm for linker histone function beyond its architectural roles in chromatin.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures
Figure 1. Multiple levels of chromatin folding
DNA compaction within the interphase nucleus occurs through a hierarchy of histone-dependent interactions, including the formation of the nucleosome core particle, strings of nucleosomes (bead-on-a-string arrangement), the chromatosome core particle and 30 nm fibres (the existence of which is debatable in vivo and which may only be relevant over short lengths of chromatin) and the association of individual fibres, which eventually produces tertiary structures.
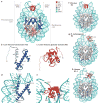
Figure 2. Structural illustration of the folded core regions of a chromatosome and representative interactions between histones and DNA
a | The crystal structure of the chromatosome core containing the globular domain of chicken H5 (H1.0; shown in red) and fold regions of core histones (H2A, H2B, H3 and H4; all colour-coded) (Protein Data Bank identifier (PDB ID): 4QLC). The globular domain sits on the dyad of the nucleosome and interacts with both linker DNAs. b | The H3 structure from part a. The structural region from α1 to α3 (in blue) is termed the histone fold, which is shared by all core histones. The dashed line represents the intrinsically disordered histone tail. c | The structure of the folded globular domain of H5 from part a. The dashed line is used to illustrate the intrinsically disordered tails. In parts b and c, N and C indicate amino termini and carboxy termini, respectively. L indicates loop regions. d | Main interactions between DNA and the core histone H3 in the nucleosome (PDB ID: 4QLC). e | Main interactions between DNA and the globular domain of H5 (PDB ID: 4QLC). Lys (K) and Arg (R) residues that presumably form electrostatic interactions with the DNA phosphates are shown in sticks and are labelled with their residue numbers. f | The on-dyad binding mode observed in the crystal structure of the mono-nucleosome bound to the globular domain of H5 (H1.0), as in part a. g | The off-dyad binding mode observed in the NMR structural model of the mono-nucleosome bound to the Drosophila melanogaster linker histone H1 (REF. 46). h | The off-dyad binding mode observed in the cryo-electron microscopy structure of the nucleosome array containing human linker histone variant H1.4 (REF. 47). The L1 loop in the globular domain is labelled to highlight the difference in the orientation of the globular domain for each binding mode. The dashed line in parts f–h indicates the nucleosome dyad.
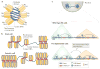
Figure 3. Roles of linker histones in chromatin folding
a | A cartoon illustration of the cryo-electron microscopy structure of the nucleosome array condensed by human linker histone H1.4 (REF. 47). The nucleosome array structure is a twisted double helix with tetra-nucleosomes as the structural unit. The globular domain of the linker histone in each nucleosome is mainly associated with one linker DNA. The globular domains of the linker histones in the nucleosomes that interact between neighbouring tetra-nucleosome units form a dimer. The linker DNA connecting the two nucleosome core particles between neighbouring tetra-nucleosome units is not associated with the globular domain of the linker histones. The tails of the linker histones in the structure are not observed. b | Nucleosomes in the presence of linker histones are likely to form chromatosomes, which are arranged in heterogeneous groups, termed ‘nucleosome clutches’, along the chromatin fibre in a cell type-specific manner: in stem cells, smaller clutches are typically observed compared with differentiated cells. This different organization of chromatosomes corresponds to differences in chromatin compaction and heterochromatization: larger clutches are associated with heterochromatin formation. c | In interphase, the chromosome is structurally organized into distinct topologically associating domains (TADs; triangles). In H1-depleted mouse embryonic stem (ES) cells, the overall genome organization into TADs is not majorly affected. In addition, interactions (black double-headed arrows) within TADs do not change. However, within gene-dense TADs, long-range inter-TAD interactions increase. In addition, new DNase hypersensitive sites (DHSs) and sites of histone H3 Lys4 mono-methylation and trimethylation (H3K4me1 and H3K4me3, respectively) are established, indicating changes in the epigenetic landscape of the cell. Part c is adapted with permission from REF. , Macmillan Publishers Limited.
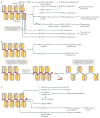
Figure 4. Biological functions of linker histones
a | Linker histones (H1) are implicated in the regulation of the epigenetic landscape of the cell by interacting with several epigenetic modifiers and by regulating their recruitment to chromatin and/or activity, which affects chromatin organization. H1 acetylated at K26 (H1K26ac) is a binding partner for the deacetylase sirtuin 1 (SIRT1), which deacetylates both core histones (H3K9ac and H4K16ac) and H1; an undefined SIRT1–H1-dependent mechanism has been further linked to hypomethylation of H3 at Lys79 (H3K79). H1 is known to recruit the histone methyltransferase Su(var)3-9, as well as DNA methyltransferases DNMT1 and DNMT3B to chromatin. Furthermore, H1 interacts with PIWI proteins and heterochromatin protein 1 (HP1), modulating histone methylation and heterochromatin formation. H1 is also a substrate for methyltransferase Polycomb repressive complex 2 (PRC2) and promotes its activity. During transcription, H1 was also shown to recruit the E3 ubiquitin ligase cullin 4A (CUL4A) and RNA polymerase II-associated factor 1 (PAF1), which is necessary for CUL4A activity. By bringing CUL4A and PAF1 together, H1 promotes CUL4A-mediated ubiquitylation of H4, which further drives methylation of core histones. H1 also repels and/or interferes with the activity of several core histone modifying enzymes, including p300/CBP-associated factor (PCAF), MOF and SET7/9. b | H1-mediated mechanisms of DNA replication control. H1 represses DNA replication at the stage of replication initiation by inhibiting the assembly of the pre-replication complex and at the stage of replication fork progression by tethering the SNF2-like ATPase protein suppressor of underreplication (SUUR). In addition, H1 undergoes S phase-dependent phosphorylation (P), which results in H1 dissociation from chromatin, leading to large-scale chromatin decondensation and the activation of origins of replication. c | The roles of H1 in DNA repair, genomic stability and DNA damage signalling. H1 is involved in DNA repair via both homologous recombination (HR) and non-homologous end joining (NHEJ) through interactions with RAD54 and with Ku86 and Ku70, respectively. H1 also facilitates ubiquitin-dependent signalling at DNA double-strand breaks: H1 is ubiquitylated by the E2 ubiquitin-conjugating enzyme UBE2N and the E3 ubiquitin ligase RNF8, and recruits the E3 ubiquitin ligase RNF168 to promote accumulation of K63-linked ubiquitin conjugates, resulting in the binding of repair factors. Chromatin compaction promoted by H1 may help to limit further DNA damage. Suppression of transposable element activity by H1 also contributes to genome stability. ICRs, imprinting control regions.
Figure 5. Biochemical activities of linker histones
a | Alternative molecular mechanisms used by linker histones (H1) to modulate the activity of chromatin. Direct biochemical interactions with H1 facilitate or inhibit chromatin binding of various structural proteins, enzymes and transcription factors; in addition, direct competition mechanisms control the mutually exclusive distribution patterns of H1 with various chromatin-interacting proteins. In general, H1-dependent chromatin compaction interferes with transcription initiation by preventing nucleosome remodelling and the binding of sequence-specific transcription factors, as well as the binding and translocation of general transcription factors. However, it has been shown that the compacted chromatin state established by H1 stimulates the activity of Polycomb repressive complex 2 (PRC2). b | Structural domains of the H1 polypeptide involved in H1 deposition, physical interactions and regulatory functions. Distinct regions within the globular domain and the carboxy-terminal domain (CTD) mediate the multiple biochemical activities of H1. CHD1, chromodomain-helicase-DNA-binding protein 1; HMG, high-mobility group; HP1, heterochromatin protein 1; MeCP2, methyl-CpG-binding protein 2; NTD, amino-terminal domain; PARP1, poly(ADP-ribose) polymerase 1; STAT, signal transducer and activator of transcription; SUUR, suppressor of underreplication.
Similar articles
- Chromatin structures condensed by linker histones.
Zhou BR, Bai Y. Zhou BR, et al. Essays Biochem. 2019 Apr 23;63(1):75-87. doi: 10.1042/EBC20180056. Print 2019 Apr 23. Essays Biochem. 2019. PMID: 31015384 Review. - Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.
Bednar J, Garcia-Saez I, Boopathi R, Cutter AR, Papai G, Reymer A, Syed SH, Lone IN, Tonchev O, Crucifix C, Menoni H, Papin C, Skoufias DA, Kurumizaka H, Lavery R, Hamiche A, Hayes JJ, Schultz P, Angelov D, Petosa C, Dimitrov S. Bednar J, et al. Mol Cell. 2017 May 4;66(3):384-397.e8. doi: 10.1016/j.molcel.2017.04.012. Mol Cell. 2017. PMID: 28475873 Free PMC article. - Linker histone defines structure and self-association behaviour of the 177 bp human chromatosome.
Wang S, Vogirala VK, Soman A, Berezhnoy NV, Liu ZB, Wong ASW, Korolev N, Su CJ, Sandin S, Nordenskiöld L. Wang S, et al. Sci Rep. 2021 Jan 11;11(1):380. doi: 10.1038/s41598-020-79654-8. Sci Rep. 2021. PMID: 33432055 Free PMC article. - Linker histones: novel insights into structure-specific recognition of the nucleosome.
Cutter AR, Hayes JJ. Cutter AR, et al. Biochem Cell Biol. 2017 Apr;95(2):171-178. doi: 10.1139/bcb-2016-0097. Epub 2016 Jun 29. Biochem Cell Biol. 2017. PMID: 28177778 Free PMC article. Review. - The Dynamic Influence of Linker Histone Saturation within the Poly-Nucleosome Array.
Woods DC, Rodríguez-Ropero F, Wereszczynski J. Woods DC, et al. J Mol Biol. 2021 May 14;433(10):166902. doi: 10.1016/j.jmb.2021.166902. Epub 2021 Mar 2. J Mol Biol. 2021. PMID: 33667509 Free PMC article.
Cited by
- Chromatin inspired bio-condensation between biomass DNA and guanosine monophosphate produces all-nucleic hydrogel as a hydrotropic drug carrier.
Sarma S, Thakur N, Varshney N, Jha HC, Sarma TK. Sarma S, et al. Commun Chem. 2024 Nov 12;7(1):261. doi: 10.1038/s42004-024-01353-6. Commun Chem. 2024. PMID: 39533097 Free PMC article. - H3K27me3 Loss in Central Nervous System Tumors: Diagnostic, Prognostic, and Therapeutic Implications.
Angelico G, Mazzucchelli M, Attanasio G, Tinnirello G, Farina J, Zanelli M, Palicelli A, Bisagni A, Barbagallo GMV, Certo F, Zizzo M, Koufopoulos N, Magro G, Caltabiano R, Broggi G. Angelico G, et al. Cancers (Basel). 2024 Oct 11;16(20):3451. doi: 10.3390/cancers16203451. Cancers (Basel). 2024. PMID: 39456545 Free PMC article. Review. - Epigenetic modulation via the C-terminal tail of H2A.Z.
Imre L, Nánási P Jr, Benhamza I, Enyedi KN, Mocsár G, Bosire R, Hegedüs É, Niaki EF, Csóti Á, Darula Z, Csősz É, Póliska S, Scholtz B, Mező G, Bacsó Z, Timmers HTM, Kusakabe M, Balázs M, Vámosi G, Ausio J, Cheung P, Tóth K, Tremethick D, Harata M, Szabó G. Imre L, et al. Nat Commun. 2024 Oct 24;15(1):9171. doi: 10.1038/s41467-024-53514-9. Nat Commun. 2024. PMID: 39448645 Free PMC article. - The Advances in the Development of Epigenetic Modifications Therapeutic Drugs Delivery Systems.
Li T, Chen Y, Li S. Li T, et al. Int J Nanomedicine. 2024 Oct 19;19:10623-10637. doi: 10.2147/IJN.S480095. eCollection 2024. Int J Nanomedicine. 2024. PMID: 39445155 Free PMC article. Review. - Navigating Latency-Inducing Viral Infections: Therapeutic Targeting and Nanoparticle Utilization.
Vasukutty A, Jang Y, Han D, Park H, Park IK. Vasukutty A, et al. Biomater Res. 2024 Oct 16;28:0078. doi: 10.34133/bmr.0078. eCollection 2024. Biomater Res. 2024. PMID: 39416703 Free PMC article. Review.
References
- Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. - PubMed
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. - PubMed
- Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. - PubMed
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM074233/GM/NIGMS NIH HHS/United States
- R01 GM093190/GM/NIGMS NIH HHS/United States
- R01 GM116143/GM/NIGMS NIH HHS/United States
- R01 GM129244/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources