Viral chimeras decrypt the role of enterovirus capsid proteins in viral tropism, acid sensitivity and optimal growth temperature - PubMed (original) (raw)
Viral chimeras decrypt the role of enterovirus capsid proteins in viral tropism, acid sensitivity and optimal growth temperature
Léna Royston et al. PLoS Pathog. 2018.
Abstract
Despite their genetic similarities, enteric and respiratory enteroviruses (EVs) have highly heterogeneous biophysical properties and cause a vast diversity of human pathologies. In vitro differences include acid sensitivity, optimal growth temperature and tissue tropism, which reflect a preferential in vivo replication in the respiratory or gastrointestinal tract and are thus key determinants of EV virulence. To investigate the underlying cause of these differences, we generated chimeras at the capsid-level between EV-D68 (a respiratory EV) and EV-D94 (an enteric EV). Although some chimeras were nonfunctional, EV-D94 with both the capsid and 2A protease or the capsid only of EV-D68 were both viable. Using this latter construct, we performed several functional assays, which indicated that capsid proteins determine acid sensitivity and tropism in cell lines and in respiratory, intestinal and neural tissues. Additionally, capsid genes were shown to also participate in determining the optimal growth temperature, since EV-D94 temperature adaptation relied on single mutations in VP1, while constructs with EV-D68 capsid could not adapt to higher temperatures. Finally, we demonstrate that EV-D68 maintains residual binding-capacity after acid-treatment despite a loss of infectivity. In contrast, non-structural rather than capsid proteins modulate the innate immune response in tissues. These unique biophysical insights expose another layer in the phenotypic diversity of one of world's most prevalent pathogens and could aid target selection for vaccine or antiviral development.
Conflict of interest statement
The authors have read the journal's policy and have the following conflicts: Song Huang and Samuel Constant are employees of Epithelix Sàrl. We confirm that this affiliation does not alter our adherence to all PLOS Pathogens policies on sharing data and materials. The other authors have no competing interests to declare.
Figures
Fig 1
A. Schematic representation of the artificially engineered chimeric EV-D68/EV-D94 viruses. Construct names are indicated on the left. EV-D68 regions are represented by white boxes; EV-D94 regions are in grey. Results obtained upon transfection and passage in HeLa cells are indicated on the right, as is the percentage of nucleotide and amino acid sequence identity between the exchanged regions. V, viable construct; X, non-viable construct; V/X, viable unfit construct. B. Non-synonymous adaptation mutations observed in viral stocks after 6 passages in HeLa cells. Sequencing was performed from nt 45 to 7344. Production of viral stocks by transfection at both 33°C and 37°C was attempted for all viruses, but only EV-D94 could be recovered at 37°C. For EV-D94, the stock subsequently used for phenotypic assessment is the one prepared at 33°C. HeLa 33 and HeLa 37: viral stocks transfected and amplified at 33°C and 37°C respectively.
Fig 2. Differential acid sensitivity of EV-D94, EV-D68 and EV-D94/D68P1.
A. Entry assay: viruses (pre-treated or not with acid) were added to cells for 2 hours at 33°C and after extensive washing, viral RNA was extracted for quantification by real-time RT-qPCR. B. Replication assay: viruses were added to cells for 1h at 33°C and after extensive washing, cells were further incubated for 24h. Viral RNA was extracted from total cell lysate and quantified. C. Binding assay: viruses were added to cells for 1 hour at 4°C to prevent entry. After extensive washing, viral RNA was extracted and quantified. D. Binding assay as in C but with cells pretreated with sialidase. For each panel, data are expressed in percentage compared to untreated controls. **P< 0.01. ***P< 0.001. ****P< 0.0001.
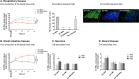
Fig 3. Differential tissue tropism of EV-D94, EV-D68 and EV-D94/D68P1.
In vitro reconstituted human tissues were inoculated with equivalent amount of EV-D94, EV-D68 and EV-D94/D68P1 and replication was assessed by RT-qPCR. A. 1st Panel: Virus production at the apical tissue side: respiratory tissues were inoculated apically and washed 3 times 4 hpi. Apical samples were collected at the indicated time point for viral RNA quantification. Residual: residual bound virus after 3 washes. 2nd Panel: Mucociliary clearance of infected respiratory tissues assessed by measuring the displacement velocity of polystyrene microbeads applied at the apical side of the tissue 5 dpi. Statistics compare infected and uninfected tissues. 3rd Panel: Immunofluorescence of respiratory tissues 5 dpi with ciliated cells stained in green, viruses in red and cell nuclei in blue. B. Small intestine tissues were infected apically and replication was quantified as for respiratory tissues. C-D. Neurons (C) and neural tissues (D) were incubated with viral suspension and viral RNA extracted from tissue lysate after inoculation and 2 or 4 days later were compared. **P< 0.01. ***P< 0.001.
Fig 4. IFN induction in respiratory tissues infected with EV-D94, EV-D68 and EV-D94/D68P1.
Respiratory tissues were inoculated apically with 108 viral particles (normalised based on RNA quantification) of the indicated virus. Apical washes were performed 4hpi and each day afterwards and tissues were lysed 3 dpi to quantify intracellular RNA (A, B and D). mRNA levels of IFN-β (A) and IFN-λ (B) in infected versus uninfected tissue were measured by RT-qPCR while IFN-λ protein levels (C) were measured in culture medium by ELISA. D. Viral loads measured 3 dpi in tissue lysates. **P< 0.01, ***P< 0.001 ****P< 0.0001.
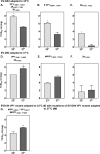
Fig 5. Temperature adaptation of EV-D94.
EV-D94 RNA transcribed from the cloned clinical isolate E210 was transfected and passaged 6 times in HeLa cells at either 33°C (“EV-D94 adapted to 33°C”, A) or 37°C (“EV-D94 adapted to 37°C, D). A. EV-D94 adapted to 33°C contains 5 non-synonymous mutations relative to the original EV-D94 clone and presents higher titers at 33°C than at 37°C. The VP1 (B) or 2A (C) mutations were introduced independently in the original infectious clone and the two derivatives retain an optimal growth at 33°C. D) EV-D94 adapted to 37°C presents 3 non synonymous mutations relative to the original EV-D94 clone and presents higher titers at 37°C than at 33°C. The VP1 (E) or 2A (F) mutations were introduced independently in the original infectious clone and only the VP1 mutated virus retains an optimal growth at 37°C. G. Mutations in VP1 of EV-D94 adapted to 37°C (E,##) were added to the VP1 mutant adapted to 33°C (B, #) and the viral titers at both temperatures were assessed. *P< 0.05, **P< 0.01.
Similar articles
- Comparison of tissue tropism and host response to enteric and respiratory enteroviruses.
Filipe IC, Tee HK, Prados J, Piuz I, Constant S, Huang S, Tapparel C. Filipe IC, et al. PLoS Pathog. 2022 Jul 5;18(7):e1010632. doi: 10.1371/journal.ppat.1010632. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35789345 Free PMC article. - Contemporary Circulating Enterovirus D68 Strains Have Acquired the Capacity for Viral Entry and Replication in Human Neuronal Cells.
Brown DM, Hixon AM, Oldfield LM, Zhang Y, Novotny M, Wang W, Das SR, Shabman RS, Tyler KL, Scheuermann RH. Brown DM, et al. mBio. 2018 Oct 16;9(5):e01954-18. doi: 10.1128/mBio.01954-18. mBio. 2018. PMID: 30327438 Free PMC article. - A Selective Bottleneck Shapes the Evolutionary Mutant Spectra of Enterovirus A71 during Viral Dissemination in Humans.
Huang SW, Huang YH, Tsai HP, Kuo PH, Wang SM, Liu CC, Wang JR. Huang SW, et al. J Virol. 2017 Nov 14;91(23):e01062-17. doi: 10.1128/JVI.01062-17. Print 2017 Dec 1. J Virol. 2017. PMID: 28931688 Free PMC article. - Global reemergence of enterovirus D68 as an important pathogen for acute respiratory infections.
Imamura T, Oshitani H. Imamura T, et al. Rev Med Virol. 2015 Mar;25(2):102-14. doi: 10.1002/rmv.1820. Epub 2014 Dec 3. Rev Med Virol. 2015. PMID: 25471236 Free PMC article. Review. - Innate Immunity Evasion by Enteroviruses: Insights into Virus-Host Interaction.
Lei X, Xiao X, Wang J. Lei X, et al. Viruses. 2016 Jan 15;8(1):22. doi: 10.3390/v8010022. Viruses. 2016. PMID: 26784219 Free PMC article. Review.
Cited by
- Evaluation of the Antiviral Activity of Sephin1 Treatment and Its Consequences on eIF2α Phosphorylation in Response to Viral Infections.
Fusade-Boyer M, Dupré G, Bessière P, Khiar S, Quentin-Froignant C, Beck C, Lecollinet S, Rameix-Welti MA, Eléouët JF, Tangy F, Lajoie B, Bertagnoli S, Vidalain PO, Gallardo F, Volmer R. Fusade-Boyer M, et al. Front Immunol. 2019 Feb 12;10:134. doi: 10.3389/fimmu.2019.00134. eCollection 2019. Front Immunol. 2019. PMID: 30809223 Free PMC article. - Comparison of tissue tropism and host response to enteric and respiratory enteroviruses.
Filipe IC, Tee HK, Prados J, Piuz I, Constant S, Huang S, Tapparel C. Filipe IC, et al. PLoS Pathog. 2022 Jul 5;18(7):e1010632. doi: 10.1371/journal.ppat.1010632. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35789345 Free PMC article. - Current Understanding of Human Enterovirus D68.
Sun J, Hu XY, Yu XF. Sun J, et al. Viruses. 2019 May 29;11(6):490. doi: 10.3390/v11060490. Viruses. 2019. PMID: 31146373 Free PMC article. Review. - The use of sialic acids as attachment factors is a common feature of _Enterovirus_-D species.
Filhol T, Mac Kain A, Joffret M-L, Jouvenet N, Caval V, Bessaud M. Filhol T, et al. J Virol. 2025 Jun 17;99(6):e0042925. doi: 10.1128/jvi.00429-25. Epub 2025 May 13. J Virol. 2025. PMID: 40358210 Free PMC article. - Neurotropism of Enterovirus D68 Isolates Is Independent of Sialic Acid and Is Not a Recently Acquired Phenotype.
Rosenfeld AB, Warren AL, Racaniello VR. Rosenfeld AB, et al. mBio. 2019 Oct 22;10(5):e02370-19. doi: 10.1128/mBio.02370-19. mBio. 2019. PMID: 31641090 Free PMC article.
References
- Picornaviridae website 2017. Available from: http://www.picornaviridae.com/enterovirus/enterovirus.htm.
- Royston L, Tapparel C. Rhinoviruses and Respiratory Enteroviruses: Not as Simple as ABC. Viruses. 2016;8(1). doi: 10.3390/v8010016 . - DOI - PMC - PubMed
- Tapparel C, Siegrist F, Petty TJ, Kaiser L. Picornavirus and enterovirus diversity with associated human diseases. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2013;14:282–93. doi: 10.1016/j.meegid.2012.10.016 . - DOI - PubMed
- Fields BN, Knipe DM, Howley PM. Fields virology. Chapter 17: Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources