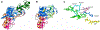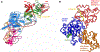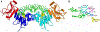Mammalian DNA methyltransferases: new discoveries and open questions - PubMed (original) (raw)
Review
. 2018 Oct 19;46(5):1191-1202.
doi: 10.1042/BST20170574. Epub 2018 Aug 28.
Affiliations
- PMID: 30154093
- PMCID: PMC6581191
- DOI: 10.1042/BST20170574
Review
Mammalian DNA methyltransferases: new discoveries and open questions
Humaira Gowher et al. Biochem Soc Trans. 2018.
Erratum in
- Correction: Mammalian DNA methyltransferases: new discoveries and open questions.
Gowher H, Jeltsch A. Gowher H, et al. Biochem Soc Trans. 2019 Jun 28;47(3):959. doi: 10.1042/BST-2017-0574C_COR. Epub 2019 Jun 24. Biochem Soc Trans. 2019. PMID: 31235548 No abstract available.
Abstract
As part of the epigenetic network, DNA methylation is a major regulator of chromatin structure and function. In mammals, it mainly occurs at palindromic CpG sites, but asymmetric methylation at non-CpG sites is also observed. Three enzymes are involved in the generation and maintenance of DNA methylation patterns. DNMT1 has high preference for hemimethylated CpG sites, and DNMT3A and DNMT3B equally methylate unmethylated and hemimethylated DNA, and also introduce non-CpG methylation. Here, we review recent observations and novel insights into the structure and function of mammalian DNMTs (DNA methyltransferases), including new structures of DNMT1 and DNMT3A, data on their mechanism, regulation by post-translational modifications and on the function of DNMTs in cells. In addition, we present news findings regarding the allosteric regulation and targeting of DNMTs by chromatin modifications and chromatin proteins. In combination, the recent publications summarized here impressively illustrate the intensity of ongoing research in this field. They provide a deeper understanding of key mechanistic properties of DNMTs, but they also document still unsolved issues, which need to be addressed in future research.
Keywords: DNA methylation; DNA methyltransferases; enzymology; molecular epigenetics.
© 2018 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
Figures
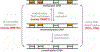
Figure 1. Cycle of DNA methylation in human cells and during development (adapted from [11]).
DNA methylation patterns are generated during development and germ cell differentiation by de novo methyltransferases and kept through DNA replication by maintenance methylation. DNA methylation can be lost through passive or active demethylation. Typically, Dnmt1 is considered to be a maintenance enzyme, whereas Dnmt3a and Dnmt3b are regarded as de novo methyltransferases (TET, ten eleven translocation enzyme; TDG, thymine–DNA glycosylase).
Figure 2. General features of DNMTs.
(A) Domain structure of the mammalian DNMT enzymes. The human DNMT1, DNMT3A, DNMT3B and DNMT3L proteins consist of 1616, 912, 853 and 387 amino acid residues, respectively. DMAPD, DNA methyltransferase-associated protein 1-interacting domain; PBD, PCNA-binding domain; NLS, nuclear localization signal; RFTD, replication foci targeting sequence (RFTS) domain; CXXC, CXXC domain; BAH1 and BAH2, bromo-adjacent homology domains 1 and 2; GKn, glycine lysine repeats; PWWP, PWWP domain; ADD, ATRX-DNMT3-DNMT3L domain (reprinted with permission from [6]). (B) Catalytic mechanism of cytosine C5 DNA methyltransferases (adapted from [12]). The catalytic glutamate residue from ENV motif (motif IV) is colored brown, the catalytic cysteine residue from PCQ motif (motif IV) is colored green, AdoMet is blue with the methyl group shown in red. The base for final proton abstraction (shown in orange) is not identified; it may be a water molecule.
Figure 3. Structures of DNMT1.
(A) Structure of the DNMT1 apoenzyme [15] ( pdb 3AV4). The RFTS domain is shown in green, CXXC in red, BAH domains in orange and purple and the catalytic domain in blue. (B) Structure of a truncated C-terminal domain of DNMT in complex with hemimethylated DNA [21] ( pdb 4DA4). SAH is shown in yellow and DNA in light green. (C) Interaction of the flipped target base (Cyt) with the catalytic residues in the active site pocket of DNMT1 (C1339, E1369 and R1315).
Figure 4. Interaction of DNMTs with chromatin modifications.
(A) Structure of the DNMT1 RFTS domain (green) in complex with the H3 peptide (shown in yellow) ubiquitinated at K18 and K23 [39] ( pdb 5WVO). The ubiquitin moieties are colored in red and light red. For illustration, the RFTS domain is overlaid with the structure of the DNMT1 apoenzyme colored and oriented as in Figure 3A. (B) Binding of the DNMT3A ADD domain to the catalytic DNMT3A domain (blue) in the DNMT3A/3L heterotetramer [55] ( pdb 4U7P and 4U7T). The ADD domain is shown in the autoinhibitory conformation (orange) and with bound peptide (yellow) in the catalytically active allosteric conformation (red).
Figure 5. Structure of DNMT3A/3L in complex with DNA [58] ( pdb 6BRR).
(A) Overview of the structure. DNMT3A subunits are shown in blue and cyan, DNMT3L subunits in red and orange. SAH is shown in yellow, DNA in green. (B) Interaction of the flipped target base (Cyt) with the catalytic residues in the active site pocket of DNMT3A.
Similar articles
- A role for LSH in facilitating DNA methylation by DNMT1 through enhancing UHRF1 chromatin association.
Han M, Li J, Cao Y, Huang Y, Li W, Zhu H, Zhao Q, Han JJ, Wu Q, Li J, Feng J, Wong J. Han M, et al. Nucleic Acids Res. 2020 Dec 2;48(21):12116-12134. doi: 10.1093/nar/gkaa1003. Nucleic Acids Res. 2020. PMID: 33170271 Free PMC article. - UHRF1 plays a role in maintaining DNA methylation in mammalian cells.
Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. Bostick M, et al. Science. 2007 Sep 21;317(5845):1760-4. doi: 10.1126/science.1147939. Epub 2007 Aug 2. Science. 2007. PMID: 17673620 - The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism.
Bashtrykov P, Jankevicius G, Jurkowska RZ, Ragozin S, Jeltsch A. Bashtrykov P, et al. J Biol Chem. 2014 Feb 14;289(7):4106-15. doi: 10.1074/jbc.M113.528893. Epub 2013 Dec 24. J Biol Chem. 2014. PMID: 24368767 Free PMC article. - Molecular enzymology of mammalian DNA methyltransferases.
Jeltsch A. Jeltsch A. Curr Top Microbiol Immunol. 2006;301:203-25. doi: 10.1007/3-540-31390-7_7. Curr Top Microbiol Immunol. 2006. PMID: 16570849 Review. - Enzymology of Mammalian DNA Methyltransferases.
Jurkowska RZ, Jeltsch A. Jurkowska RZ, et al. Adv Exp Med Biol. 2016;945:87-122. doi: 10.1007/978-3-319-43624-1_5. Adv Exp Med Biol. 2016. PMID: 27826836 Review.
Cited by
- Using human disease mutations to understand de novo DNA methyltransferase function.
Rolls W, Wilson MD, Sproul D. Rolls W, et al. Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review. - Allele-specific DNA demethylation editing leads to stable upregulation of allele-specific gene expression.
Rajaram N, Benzler K, Bashtrykov P, Jeltsch A. Rajaram N, et al. iScience. 2024 Sep 23;27(10):111007. doi: 10.1016/j.isci.2024.111007. eCollection 2024 Oct 18. iScience. 2024. PMID: 39429790 Free PMC article. - Effects of Cohabitation on Neurodevelopmental Outcomes in Rats Discordant for Neonatal Exposure to Sevoflurane.
Ju LS, Morey T, Gravenstein N, Setlow B, Seubert CN, Martynyuk AE. Ju LS, et al. Biol Psychiatry Glob Open Sci. 2024 Jul 15;4(6):100359. doi: 10.1016/j.bpsgos.2024.100359. eCollection 2024 Nov. Biol Psychiatry Glob Open Sci. 2024. PMID: 39282654 Free PMC article. - The association between cumulative exposure to PM2.5 and DNA methylation measured using methyl-capture sequencing among COPD patients.
Ji HW, Kang J, Kim HC, Jung J, Lee SJ, Jung JY, Lee SW. Ji HW, et al. Respir Res. 2024 Sep 9;25(1):335. doi: 10.1186/s12931-024-02955-3. Respir Res. 2024. PMID: 39251997 Free PMC article. - DNA methylation in mammalian development and disease.
Smith ZD, Hetzel S, Meissner A. Smith ZD, et al. Nat Rev Genet. 2024 Aug 12. doi: 10.1038/s41576-024-00760-8. Online ahead of print. Nat Rev Genet. 2024. PMID: 39134824 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials