ArrayExpress update - from bulk to single-cell expression data - PubMed (original) (raw)
. 2019 Jan 8;47(D1):D711-D715.
doi: 10.1093/nar/gky964.
Anja Füllgrabe 1, Nancy George 1, Haider Iqbal 1, Laura Huerta 1, Ahmed Ali 1, Catherine Snow 1, Nuno A Fonseca 2, Robert Petryszak 1, Irene Papatheodorou 1, Ugis Sarkans 1, Alvis Brazma 1
Affiliations
- PMID: 30357387
- PMCID: PMC6323929
- DOI: 10.1093/nar/gky964
ArrayExpress update - from bulk to single-cell expression data
Awais Athar et al. Nucleic Acids Res. 2019.
Abstract
ArrayExpress (https://www.ebi.ac.uk/arrayexpress) is an archive of functional genomics data from a variety of technologies assaying functional modalities of a genome, such as gene expression or promoter occupancy. The number of experiments based on sequencing technologies, in particular RNA-seq experiments, has been increasing over the last few years and submissions of sequencing data have overtaken microarray experiments in the last 12 months. Additionally, there is a significant increase in experiments investigating single cells, rather than bulk samples, known as single-cell RNA-seq. To accommodate these trends, we have substantially changed our submission tool Annotare which, along with raw and processed data, collects all metadata necessary to interpret these experiments. Selected datasets are re-processed and loaded into our sister resource, the value-added Expression Atlas (and its component Single Cell Expression Atlas), which not only enables users to interpret the data easily but also serves as a test for data quality. With an increasing number of studies that combine different assay modalities (multi-omics experiments), a new more general archival resource the BioStudies Database has been developed, which will eventually supersede ArrayExpress. Data submissions will continue unchanged; all existing ArrayExpress data will be incorporated into BioStudies and the existing accession numbers and application programming interfaces will be maintained.
Figures
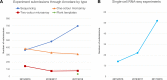
Figure 1.
(A) Experiment submissions to ArrayExpress via Annotare during the last 3 years by experiment type. Data from September 2017 to August 2018 include 34 submissions using the newly introduced plant templates (27 plant sequencing, 5 plant one-colour microarray and 2 plant two-colour microarray experiments). (B) Number of experiments in ArrayExpress (public or private) with experiment type ‘RNA-seq of coding RNA from single cells’ by submission year.
Figure 2.
Annotare submission set-up and template selection. First, the user chooses the template type e.g. ‘Plant - high-throughput sequencing’. Then, the study design can be selected from a list of suggestions. Based on these choices, the relevant sample attributes and experimental variables are preselected.
Figure 3.
The updated Annotare interface featuring an uncoupled file upload panel, simplified navigation, improved validation error reporting and a new protocol entry panel.
Similar articles
- From ArrayExpress to BioStudies.
Sarkans U, Füllgrabe A, Ali A, Athar A, Behrangi E, Diaz N, Fexova S, George N, Iqbal H, Kurri S, Munoz J, Rada J, Papatheodorou I, Brazma A. Sarkans U, et al. Nucleic Acids Res. 2021 Jan 8;49(D1):D1502-D1506. doi: 10.1093/nar/gkaa1062. Nucleic Acids Res. 2021. PMID: 33211879 Free PMC article. - ArrayExpress update--simplifying data submissions.
Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, Dylag M, Kurbatova N, Brandizi M, Burdett T, Megy K, Pilicheva E, Rustici G, Tikhonov A, Parkinson H, Petryszak R, Sarkans U, Brazma A. Kolesnikov N, et al. Nucleic Acids Res. 2015 Jan;43(Database issue):D1113-6. doi: 10.1093/nar/gku1057. Epub 2014 Oct 31. Nucleic Acids Res. 2015. PMID: 25361974 Free PMC article. - ArrayExpress update--from an archive of functional genomics experiments to the atlas of gene expression.
Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, Holloway E, Lukk M, Malone J, Mani R, Pilicheva E, Rayner TF, Rezwan F, Sharma A, Williams E, Bradley XZ, Adamusiak T, Brandizi M, Burdett T, Coulson R, Krestyaninova M, Kurnosov P, Maguire E, Neogi SG, Rocca-Serra P, Sansone SA, Sklyar N, Zhao M, Sarkans U, Brazma A. Parkinson H, et al. Nucleic Acids Res. 2009 Jan;37(Database issue):D868-72. doi: 10.1093/nar/gkn889. Epub 2008 Nov 10. Nucleic Acids Res. 2009. PMID: 19015125 Free PMC article. - Data storage and analysis in ArrayExpress.
Brazma A, Kapushesky M, Parkinson H, Sarkans U, Shojatalab M. Brazma A, et al. Methods Enzymol. 2006;411:370-86. doi: 10.1016/S0076-6879(06)11020-4. Methods Enzymol. 2006. PMID: 16939801 Review. - Gene expression omnibus: microarray data storage, submission, retrieval, and analysis.
Barrett T, Edgar R. Barrett T, et al. Methods Enzymol. 2006;411:352-69. doi: 10.1016/S0076-6879(06)11019-8. Methods Enzymol. 2006. PMID: 16939800 Free PMC article. Review.
Cited by
- HUSCH: an integrated single-cell transcriptome atlas for human tissue gene expression visualization and analyses.
Shi X, Yu Z, Ren P, Dong X, Ding X, Song J, Zhang J, Li T, Wang C. Shi X, et al. Nucleic Acids Res. 2023 Jan 6;51(D1):D1029-D1037. doi: 10.1093/nar/gkac1001. Nucleic Acids Res. 2023. PMID: 36318258 Free PMC article. - Feature importance network reveals novel functional relationships between biological features in Arabidopsis thaliana.
Ng JWX, Chua SK, Mutwil M. Ng JWX, et al. Front Plant Sci. 2022 Sep 23;13:944992. doi: 10.3389/fpls.2022.944992. eCollection 2022. Front Plant Sci. 2022. PMID: 36212273 Free PMC article. - Meta-Analysis of Oxidative Transcriptomes in Insects.
Bono H. Bono H. Antioxidants (Basel). 2021 Feb 25;10(3):345. doi: 10.3390/antiox10030345. Antioxidants (Basel). 2021. PMID: 33669076 Free PMC article. - In-Silico Identification of Novel Pharmacological Synergisms: The Trabectedin Case.
Mannarino L, Ravasio N, D'Incalci M, Marchini S, Masseroli M. Mannarino L, et al. Int J Mol Sci. 2024 Feb 8;25(4):2059. doi: 10.3390/ijms25042059. Int J Mol Sci. 2024. PMID: 38396735 Free PMC article. - Comprehensive RNA dataset of tissue and plasma from patients with esophageal cancer or precursor lesions.
Schoofs K, Philippron A, Avila Cobos F, Koster J, Lefever S, Anckaert J, De Looze D, Vandesompele J, Pattyn P, De Preter K. Schoofs K, et al. Sci Data. 2022 Mar 14;9(1):86. doi: 10.1038/s41597-022-01176-x. Sci Data. 2022. PMID: 35288573 Free PMC article.
References
- Parkinson H., Sarkans U., Kolesnikov N., Abeygunawardena N., Burdett T., Dylag M., Emam I., Farne A., Hastings E., Holloway E. et al. . ArrayExpress update–an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011; 39:D1002–D1004. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources