Evaluating gut microbiota profiles from archived fecal samples - PubMed (original) (raw)
Evaluating gut microbiota profiles from archived fecal samples
Trine B Rounge et al. BMC Gastroenterol. 2018.
Abstract
Background: Associations between colorectal cancer and microbiota have been identified. Archived fecal samples might be valuable sample sources for investigating causality in carcinogenesis and biomarkers discovery due to the potential of performing longitudinal studies. However, the quality, quantity and stability of the gut microbiota in these fecal samples must be assessed prior to such studies. We evaluated i) cross-contamination during analysis for fecal blood and ii) evaporation in stored perforated fecal immunochemical tests (iFOBT) samples, iii) temperature stability as well as iv) comparison of the gut microbiota diversity and composition in archived, iFOBT and fresh fecal samples in order to assess feasibility of large scale microbiota studies.
Methods: The microbiota profiles were obtained by sequencing the V3-V4 region of 16S rDNA gene.
Results: The iFOBT does not introduce any cross-sample contamination detectable by qPCR. Neither could we detect evaporation during freeze-thaw cycle of perforated iFOBT samples. Our results confirm room temperature stability of the gut microbiome. Diverse microbial profiles were achieved in 100% of fresh, 81% of long-term archived and 96% of iFOBT samples. Microbial diversity and composition were comparable between fresh and iFOBT samples, however, diversity differed significantly between long-term archived, fresh and iFOBT samples.
Conclusion: Our data showed that it is feasible to exploit archived fecal sample sets originally collected for testing of fecal blood. The advantages of using these sample sets for microbial biomarker discovery and longitudinal observational studies are the availability of high-quality diagnostic and follow-up data. However, care must be taken when microbiota are profiled in long-term archived fecal samples.
Keywords: Archived samples; Diversity; Fecal immunochemical tests; Fecal samples; Microbiota; Storage.
Conflict of interest statement
Ethics approval and consent to participate
The collection of iFOBT and archived samples was approved by the Regional Committees for Medical and Health Research Ethics in South-Eastern Norway (2011/1272 and 2010/3087 A, respectively). An additional ethical approval for the feasibility study was not required since all samples were anonymized and the purpose of the study was method related, as stated by the Norwegian regional ethics committee (Regional Committees for Medical and Health Research Ethics in South-Eastern Norway, ref. 2015/9).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests, with the exception of GH received payment from Amgen Norway for giving a lecture at a medical conference in 2017.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
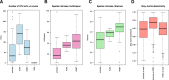
Fig. 1
Alpha and beta diversity in iFOBT, fresh and archived samples. a Shows a boxplot of the number of observed OTUs in each sample group. b and c Shows boxplots of the Inverse Simpson index (b) and Shannon (c) index in fecal immunochemical tests (iFOBT samples, fresh fecal samples and fecal samples archived for approximately 16 years. The indexes are based on rarefied OTU data to minimize the impacts of uneven sampling. The Bray-Curtis dissimilarity index for comparisons of groups are shown (d)
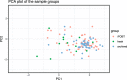
Fig. 2
Principal component plot of the community composition of iFOBT, archived and fresh sample. The first and second principal components of the community composition of iFOBT samples from a screening trial in Norway (BCSN), archived samples from the NORCCAP cohort stored for about 16 years and fresh samples
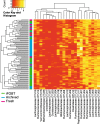
Fig. 3
Clustering of archived, fresh and fecal immunochemical tests (iFOBT) fecal samples. Heatmap of the log transformed OTU table for all samples produced by hierarchical clustering and Euclidean distance. Only OTUs with a log sum > 20 were illustrated. iFOBT samples are marked in green, archived fecal samples stored for 14 to 16 years at − 30 °C are marked in blue and fresh samples are marked in pink. All samples are from presumably healthy individuals
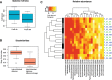
Fig. 4
Microbiota profiles in fresh samples frozen directly and frozen after 48 h in room temperature. a Boxplot of Inverse Simpson alpha-diversity index in samples frozen directly and frozen after 48 h. b Bray-Curtis dissimilarities between samples from the same individuals with different storage conditions, and between different individuals regardless of room temperature storage. c Heatmap of the log transformed OTU table from paired fecal samples from 8 presumably healthy individuals (serial number 01 to 10). One part of the samples was directly frozen (0 h, marked with green text) and the other part of the samples was frozen after 48 h in room temperature (48 h, marked with blue text). Hierarchical clustering and Euclidean distance produced the clustering showing higher inter-person variability than intra-person variability. Only OTUs with a log sum > 2 were illustrated
Similar articles
- Collection of non-meconium stool on fecal occult blood cards is an effective method for fecal microbiota studies in infants.
Wong WSW, Clemency N, Klein E, Provenzano M, Iyer R, Niederhuber JE, Hourigan SK. Wong WSW, et al. Microbiome. 2017 Sep 5;5(1):114. doi: 10.1186/s40168-017-0333-z. Microbiome. 2017. PMID: 28870234 Free PMC article. - Assessment of the impact of different fecal storage protocols on the microbiota diversity and composition: a pilot study.
Moossavi S, Engen PA, Ghanbari R, Green SJ, Naqib A, Bishehsari F, Merat S, Poustchi H, Keshavarzian A, Malekzadeh R. Moossavi S, et al. BMC Microbiol. 2019 Jun 28;19(1):145. doi: 10.1186/s12866-019-1519-2. BMC Microbiol. 2019. PMID: 31253096 Free PMC article. - High-throughput DNA extraction strategy for fecal microbiome studies.
Isokääntä H, Tomnikov N, Vanhatalo S, Munukka E, Huovinen P, Hakanen AJ, Kallonen T. Isokääntä H, et al. Microbiol Spectr. 2024 Jun 4;12(6):e0293223. doi: 10.1128/spectrum.02932-23. Epub 2024 May 15. Microbiol Spectr. 2024. PMID: 38747618 Free PMC article. - Effects of Stool Sample Preservation Methods on Gut Microbiota Biodiversity: New Original Data and Systematic Review with Meta-Analysis.
Li XM, Shi X, Yao Y, Shen YC, Wu XL, Cai T, Liang LX, Wang F. Li XM, et al. Microbiol Spectr. 2023 Jun 15;11(3):e0429722. doi: 10.1128/spectrum.04297-22. Epub 2023 Apr 24. Microbiol Spectr. 2023. PMID: 37093040 Free PMC article. - Systematic review: Gut microbiota in fecal samples and detection of colorectal neoplasms.
Amitay EL, Krilaviciute A, Brenner H. Amitay EL, et al. Gut Microbes. 2018 Jul 4;9(4):293-307. doi: 10.1080/19490976.2018.1445957. Epub 2018 May 15. Gut Microbes. 2018. PMID: 29543545 Free PMC article.
Cited by
- Profiling small RNAs in fecal immunochemical tests: is it possible?
Birkeland E, Ferrero G, Pardini B, Umu SU, Tarallo S, Bulfamante S, Hoff G, Senore C, Rounge TB, Naccarati A. Birkeland E, et al. Mol Cancer. 2023 Oct 3;22(1):161. doi: 10.1186/s12943-023-01869-w. Mol Cancer. 2023. PMID: 37789383 Free PMC article. - Reusing a prepaid health plan's fecal immunochemical tests for microbiome associations with colorectal adenoma.
Goedert JJ, Wu Z, Yonehara CH, Frankland TB, Sinha R, Jones GS, Wan Y, Ravel J, Zhao N, Honda SA. Goedert JJ, et al. Sci Rep. 2022 Aug 31;12(1):14801. doi: 10.1038/s41598-022-18870-w. Sci Rep. 2022. PMID: 36045142 Free PMC article. - Exploring the gut DNA virome in fecal immunochemical test stool samples reveals associations with lifestyle in a large population-based study.
Istvan P, Birkeland E, Avershina E, Kværner AS, Bemanian V, Pardini B, Tarallo S, de Vos WM, Rognes T, Berstad P, Rounge TB. Istvan P, et al. Nat Commun. 2024 Feb 29;15(1):1791. doi: 10.1038/s41467-024-46033-0. Nat Commun. 2024. PMID: 38424056 Free PMC article. - The CRCbiome study: a large prospective cohort study examining the role of lifestyle and the gut microbiome in colorectal cancer screening participants.
Kværner AS, Birkeland E, Bucher-Johannessen C, Vinberg E, Nordby JI, Kangas H, Bemanian V, Ellonen P, Botteri E, Natvig E, Rognes T, Hovig E, Lyle R, Ambur OH, de Vos WM, Bultman S, Hjartåker A, Landberg R, Song M, Blix HS, Ursin G, Randel KR, de Lange T, Hoff G, Holme Ø, Berstad P, Rounge TB. Kværner AS, et al. BMC Cancer. 2021 Aug 18;21(1):930. doi: 10.1186/s12885-021-08640-8. BMC Cancer. 2021. PMID: 34407780 Free PMC article. Clinical Trial. - Different living environments drive deterministic microbial community assemblages in the gut of Alpine musk deer (Moschus chrysogaster).
Zhang Z, Ding M, Sun Y, Khattak RH, Chen J, Teng L, Liu Z. Zhang Z, et al. Front Microbiol. 2023 Jan 13;13:1108405. doi: 10.3389/fmicb.2022.1108405. eCollection 2022. Front Microbiol. 2023. PMID: 36713154 Free PMC article.
References
- Colorectal cancer estimated incidence, mortality and prevalence worldwide in 2012. [http://globocan.iarc.fr/old/FactSheets/cancers/colorectal-new.asp].
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical