HistoQC: An Open-Source Quality Control Tool for Digital Pathology Slides - PubMed (original) (raw)
HistoQC: An Open-Source Quality Control Tool for Digital Pathology Slides
Andrew Janowczyk et al. JCO Clin Cancer Inform. 2019 Apr.
Abstract
Purpose: Digital pathology (DP), referring to the digitization of tissue slides, is beginning to change the landscape of clinical diagnostic workflows and has engendered active research within the area of computational pathology. One of the challenges in DP is the presence of artefacts and batch effects, unintentionally introduced during both routine slide preparation (eg, staining, tissue folding) and digitization (eg, blurriness, variations in contrast and hue). Manual review of glass and digital slides is laborious, qualitative, and subject to intra- and inter-reader variability. Therefore, there is a critical need for a reproducible automated approach of precisely localizing artefacts to identify slides that need to be reproduced or regions that should be avoided during computational analysis.
Methods: Here we present HistoQC, a tool for rapidly performing quality control to not only identify and delineate artefacts but also discover cohort-level outliers (eg, slides stained darker or lighter than others in the cohort). This open-source tool employs a combination of image metrics (eg, color histograms, brightness, contrast), features (eg, edge detectors), and supervised classifiers (eg, pen detection) to identify artefact-free regions on digitized slides. These regions and metrics are presented to the user via an interactive graphical user interface, facilitating artefact detection through real-time visualization and filtering. These same metrics afford users the opportunity to explicitly define acceptable tolerances for their workflows.
Results: The output of HistoQC on 450 slides from The Cancer Genome Atlas was reviewed by two pathologists and found to be suitable for computational analysis more than 95% of the time.
Conclusion: These results suggest that HistoQC could provide an automated, quantifiable, quality control process for identifying artefacts and measuring slide quality, in turn helping to improve both the repeatability and robustness of DP workflows.
Conflict of interest statement
Andrew R. Janowczyk
Consulting or Advisory Role: Merck
Hannah Gilmore
Travel, Accommodations, Expenses: Sectra
Michael Feldman
Consulting or Advisory Role: Philips Healthcare
Travel, Accommodations, Expenses: Philips Healthcare
Anant Madabhushi
Leadership: Inspirata
Stock and Other Ownership Interests: Inspirata, Elucid Bioimaging
Honoraria: AstraZeneca, Inspirata
Consulting or Advisory Role: Inspirata, AstraZeneca, Merck
Research Funding: Inspirata (Inst), Philips Healthcare (Inst)
Patents, Royalties, Other Intellectual Property: Intellectual property licensed by Inspirata (Inst); intellectual property licensed by Elucid Bioimaging (Inst)
No other potential conflicts of interest were reported.
Figures
FIG 1.
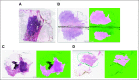
Original images juxtaposed with corresponding results from HistoQC (fuchsia indicating acceptable tissue): images identified as having (A) a significant air bubble artefact requiring removal from experimental cohort, (B) blurry tissue near a coverslip crack, (C) folded tissue, and (D) pen markings correctly identified as regions to be avoided.
FIG 2.
(A) HistoQC user interface showing table of HistoQC-produced metrics with sortable columns (green arrow), parallel coordinate plot (red box; additional details in Fig 3), and thumbnail images of the cohort alongside HistoQC overlay output indicating artefact-free regions (blue arrow). (B) Selecting a single image highlights the appropriate line in the parallel coordinate graph and shows the series of outputs produced by the modules of the pipeline, allowing for more detailed subsequent review. (C) Double clicking on any image brings up a higher-resolution version with dynamic zoom, allowing for fine-tuned inspection of potential artefacts. Hist, histogram; MPP, microns per pixel; MSE, mean squared error.
FIG 3.
(A) Higher magnification of the parallel coordinate plot bounded by the red box in Figure 2. The different _y_-axes correspond to different image metrics determined by HistoQC and may have their own ranges and scales. Each horizontally oriented line, in either gray or blue, represents a whole-slide image (WSI) analyzed by HistoQC. By examining the convergence or divergence of each horizontal WSI line with respect to the rest of the images in the cohort, batch effects and outliers can be more easily visually identified. In the example illustrated, the user has interactively drawn a gray box (red arrow) to select images with a grayscale intensity value between 0.4 and 0.5, resulting in all lines that do not meet this criterion turning gray. This image selection mechanism alters the visibility of slides in both the table (Fig 2 green arrow) and the thumbnails (Fig 2 blue arrow). Furthermore, the user can dynamically drag or extend the gray box upward and downward to update the visible slides in real time. As a result, images with (B) low and (C) high grayscale brightness values are easier to identify and review.
Similar articles
- Assessment of a computerized quantitative quality control tool for whole slide images of kidney biopsies.
Chen Y, Zee J, Smith A, Jayapandian C, Hodgin J, Howell D, Palmer M, Thomas D, Cassol C, Farris AB 3rd, Perkinson K, Madabhushi A, Barisoni L, Janowczyk A. Chen Y, et al. J Pathol. 2021 Mar;253(3):268-278. doi: 10.1002/path.5590. Epub 2021 Jan 5. J Pathol. 2021. PMID: 33197281 Free PMC article. - Applications and challenges of digital pathology and whole slide imaging.
Higgins C. Higgins C. Biotech Histochem. 2015 Jul;90(5):341-7. doi: 10.3109/10520295.2015.1044566. Epub 2015 May 15. Biotech Histochem. 2015. PMID: 25978139 Review. - Whole slide imaging equivalency and efficiency study: experience at a large academic center.
Hanna MG, Reuter VE, Hameed MR, Tan LK, Chiang S, Sigel C, Hollmann T, Giri D, Samboy J, Moradel C, Rosado A, Otilano JR 3rd, England C, Corsale L, Stamelos E, Yagi Y, Schüffler PJ, Fuchs T, Klimstra DS, Sirintrapun SJ. Hanna MG, et al. Mod Pathol. 2019 Jul;32(7):916-928. doi: 10.1038/s41379-019-0205-0. Epub 2019 Feb 18. Mod Pathol. 2019. PMID: 30778169 - iPathology cockpit diagnostic station: validation according to College of American Pathologists Pathology and Laboratory Quality Center recommendation at the Hospital Trust and University of Verona.
Brunelli M, Beccari S, Colombari R, Gobbo S, Giobelli L, Pellegrini A, Chilosi M, Lunardi M, Martignoni G, Scarpa A, Eccher A. Brunelli M, et al. Diagn Pathol. 2014;9 Suppl 1(Suppl 1):S12. doi: 10.1186/1746-1596-9-S1-S12. Epub 2014 Dec 19. Diagn Pathol. 2014. PMID: 25565219 Free PMC article. - Digital Pathology for Better Clinical Practice.
Hijazi A, Bifulco C, Baldin P, Galon J. Hijazi A, et al. Cancers (Basel). 2024 Apr 26;16(9):1686. doi: 10.3390/cancers16091686. Cancers (Basel). 2024. PMID: 38730638 Free PMC article. Review.
Cited by
- AI drives the assessment of lung cancer microenvironment composition.
Gallo E, Guardiani D, Betti M, Arteni BAM, Di Martino S, Baldinelli S, Daralioti T, Merenda E, Ascione A, Visca P, Pescarmona E, Lavitrano M, Nisticò P, Ciliberto G, Pallocca M. Gallo E, et al. J Pathol Inform. 2024 Sep 30;15:100400. doi: 10.1016/j.jpi.2024.100400. eCollection 2024 Dec. J Pathol Inform. 2024. PMID: 39469280 Free PMC article. - Equipping computational pathology systems with artifact processing pipelines: a showcase for computation and performance trade-offs.
Kanwal N, Khoraminia F, Kiraz U, Mosquera-Zamudio A, Monteagudo C, Janssen EAM, Zuiverloon TCM, Rong C, Engan K. Kanwal N, et al. BMC Med Inform Decis Mak. 2024 Oct 7;24(1):288. doi: 10.1186/s12911-024-02676-z. BMC Med Inform Decis Mak. 2024. PMID: 39375719 Free PMC article. - A novel computational pathology approach for identifying gene signatures prognostic of disease-free survival for papillary thyroid carcinomas.
Monabbati S, Khalighi S, Fu P, Shi Q, Asa SL, Madabhushi A. Monabbati S, et al. Eur J Cancer. 2024 Nov;212:114326. doi: 10.1016/j.ejca.2024.114326. Epub 2024 Sep 17. Eur J Cancer. 2024. PMID: 39307037 - From whole-slide image to biomarker prediction: end-to-end weakly supervised deep learning in computational pathology.
El Nahhas OSM, van Treeck M, Wölflein G, Unger M, Ligero M, Lenz T, Wagner SJ, Hewitt KJ, Khader F, Foersch S, Truhn D, Kather JN. El Nahhas OSM, et al. Nat Protoc. 2024 Sep 16. doi: 10.1038/s41596-024-01047-2. Online ahead of print. Nat Protoc. 2024. PMID: 39285224 Review. - Optimization of whole slide imaging scan settings for computer vision using human lung cancer tissue.
Geubbelmans M, Claes J, Nijsten K, Gervois P, Appeltans S, Martens S, Wolfs E, Thomeer M, Valkenborg D, Faes C. Geubbelmans M, et al. PLoS One. 2024 Sep 9;19(9):e0309740. doi: 10.1371/journal.pone.0309740. eCollection 2024. PLoS One. 2024. PMID: 39250489 Free PMC article.
References
- Doyle S, Rodriguez C, Madabhushi A, et al: Detecting prostatic adenocarcinoma from digitized histology using a multi-scale hierarchical classification approach. Conf Proc IEEE Eng Med Biol Soc 1:4759-4762, 2006. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 CA216579/CA/NCI NIH HHS/United States
- C06 RR012463/RR/NCRR NIH HHS/United States
- U24 CA199374/CA/NCI NIH HHS/United States
- I01 BX004121/BX/BLRD VA/United States
- T32 DK007470/DK/NIDDK NIH HHS/United States
- R01 CA220581/CA/NCI NIH HHS/United States
- R01 CA202752/CA/NCI NIH HHS/United States
- R01 CA208236/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources