A conserved role of the insulin-like signaling pathway in diet-dependent uric acid pathologies in Drosophila melanogaster - PubMed (original) (raw)
. 2019 Aug 15;15(8):e1008318.
doi: 10.1371/journal.pgen.1008318. eCollection 2019 Aug.
Tyler A Hilsabeck 1 2, Kenneth A Wilson 1 2, Amit Sharma 1, Neelanjan Bose 1, Deanna J Brackman 3, Jennifer N Beck 1, Ling Chen 4, Mark A Watson 1, David W Killilea 5, Sunita Ho 4, Arnold Kahn 1, Kathleen Giacomini 3, Marshall L Stoller 6, Thomas Chi 6, Pankaj Kapahi 1
Affiliations
- PMID: 31415568
- PMCID: PMC6695094
- DOI: 10.1371/journal.pgen.1008318
A conserved role of the insulin-like signaling pathway in diet-dependent uric acid pathologies in Drosophila melanogaster
Sven Lang et al. PLoS Genet. 2019.
Abstract
Elevated uric acid (UA) is a key risk factor for many disorders, including metabolic syndrome, gout and kidney stones. Despite frequent occurrence of these disorders, the genetic pathways influencing UA metabolism and the association with disease remain poorly understood. In humans, elevated UA levels resulted from the loss of the of the urate oxidase (Uro) gene around 15 million years ago. Therefore, we established a Drosophila melanogaster model with reduced expression of the orthologous Uro gene to study the pathogenesis arising from elevated UA. Reduced Uro expression in Drosophila resulted in elevated UA levels, accumulation of concretions in the excretory system, and shortening of lifespan when reared on diets containing high levels of yeast extract. Furthermore, high levels of dietary purines, but not protein or sugar, were sufficient to produce the same effects of shortened lifespan and concretion formation in the Drosophila model. The insulin-like signaling (ILS) pathway has been shown to respond to changes in nutrient status in several species. We observed that genetic suppression of ILS genes reduced both UA levels and concretion load in flies fed high levels of yeast extract. Further support for the role of the ILS pathway in modulating UA metabolism stems from a human candidate gene study identifying SNPs in the ILS genes AKT2 and FOXO3 being associated with serum UA levels or gout. Additionally, inhibition of the NADPH oxidase (NOX) gene rescued the reduced lifespan and concretion phenotypes in Uro knockdown flies. Thus, components of the ILS pathway and the downstream protein NOX represent potential therapeutic targets for treating UA associated pathologies, including gout and kidney stones, as well as extending human healthspan.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
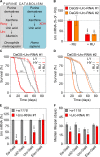
Fig 1. Uro knockdown attenuates lifespan on a high yeast diet.
(A) Schematic comparison of the purine catabolism of Drosophila melanogaster and humans with relevant enzymes in red and metabolites in black. (B) Uro mRNA levels were determined in presence and absence of RU486 by qRT-PCR. Progeny of crosses combining the daughterless gene switch (DaGS) driver with one of the two different Uro targeting RNAi constructs (_Uro_-RNAi #1, _Uro_-RNAi #2) were fed diets without RU486 (- RU) or with RU486 (+ RU). For each cross Uro mRNA value of the - RU diet was set as 100%. (C, D) Kaplan-Meier survival curves of DaGS>_Uro_-RNAi #1 (C) and DaGS>_Uro_-RNAi #2 (D) flies fed diets containing a low (LY; 0.5%) or high yeast extract content (HY; 5%) +/- RU. (E) Relative Uro mRNA levels of different Gal4 driver lines (Da-GAL4, c42-GAL4, Uro-GAL4, Elav-GAL4) crossed with the background control w1118 or the _Uro_-RNAi #1. For each cross with w1118 the Uro mRNA value was set to 100%. (F) Average median lifespan of flies from (E) fed the HY diet. The average median lifespan was deduced from multiple survival curves and is defined as the time point in days when 50% of the population is alive. Error bars represent the standard error (SE). With the exception of the LY conditions in (D) all experiments were run in independent biological repeats. Supporting information is given in S1 Fig.
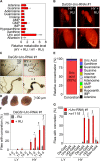
Fig 2. Uro knockdown enhances UA concretion formation on a high yeast diet.
(A) Mass spectrometric analysis of purine metabolite concentrations of DaGS>_Uro_-RNAi #1 fly homogenates comparing siblings fed the HY + RU and HY - RU diet for 14 days. (B) Micro-CT images of intact flies from (A). Fly length is ~2 mm. (C) Guts with the attached Malpighian tubules were dissected from flies in (B). Black arrowheads point to solid aggregates (concretions) in the hindgut region. (D) Light microscopic pictures of concretions extracted from the hindgut lumen of DaGS>_Uro_-RNAi #1 flies fed the HY diet + RU for 14 days. (E) Metabolomic analysis of the same purine metabolites as in (A) of extracted concretions from Uro knockdown flies. (F) The extent and kinetic of concretion formation in DaGS>_Uro_-RNAi #1 flies fed the LY or HY diet +/- RU for 4 to 14 days was determined by microscopic analysis after dissection of the hindgut. (G) Concretion formation of flies from Fig 1E fed the LY or HY diet for 14 days was determined by microscopic analysis after dissection of the hindgut. Error bars represent the SE of multiple biological repeats. Further supporting information is shown in S2 Fig.
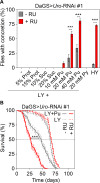
Fig 3. Dietary purine mediates lifespan attenuation and UA concretion formation.
(A) Concretion formation of DaGS>_Uro_-RNAi #1 flies reared on a HY or LY diet for 14 days. Where indicated, the latter was supplemented with increasing concentrations of protein (Prot), sucrose (Suc), purine (Pu) or pyrimidine (Py). Each food was generated in a - RU and + RU version to compare concretion formation of isogenic control and Uro knockdown siblings, respectively. (B) Survival curve of DaGS>_Uro_-RNAi #1 flies fed the LY diet without or with 40 mM purine supplementation (+ Pu) in presence or absence of RU.
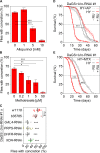
Fig 4. Modulation of purine homeostasis rescues the lifespan attenuation and UA concretion formation of Uro knockdown flies.
(A, B) Concretion formation of DaGS>_Uro_-RNAi #1 flies after 14 days of feeding a HY + RU diet supplemented with the indicated concentration of allopurinol (A) or methotrexate (B). (C) The recombinant DaGS::_Uro_-RNAi #1 line was crossed to w1118 (no additional UAS-locus), b35785 (carrying a no target UAS-mCherry-RNAi), or active UAS-RNAi lines targeting the transcription factor GAL4, PRPP synthetase (PRPS), dihydrofolate reductase (DHFR), or xanthine dehydrogenase (XDH). To measure concretion formation the flies were fed the HY + RU diet for 14 days prior to dissection. (D) Survival curve of DaGS>_Uro_-RNAi #1 flies fed the HY diet without or with 5 mM AP (+AP) supplementation in presence or absence of RU. (E) As in (D), but 5 μM MTX (+MTX) supplementation. Error bars represent the SE of multiple biological repeats. Supporting data is shown in S3 Fig.
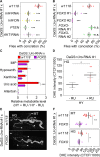
Fig 5. FoxO over-expression inhibits concretion formation by reducing UA levels and ROS formation.
(A, B) The recombinant DaGS::_Uro_-RNAi #1 line was crossed to w1118 (no additional UAS-locus), or strains with active UAS-transgenes triggering either inhibition of the insulin-like receptor (_InR_-RNAi, _InR_-DN (dominant negative)), AKT kinase (_AKT_-RNAi), or the transcription factor FoxO (_FOXO_-RNAi #1, _FOXO_-RNAi #2), or triggering over-expression of PTEN, or FOXO (FOXO #1, FOXO #2). Concretion formation was checked after 14 days of feeding the HY + RU diet. (C) Mass spectrometric analysis of purine metabolite concentrations of fly homogenates from DaGS::_Uro_-RNAi #1 crossed to w1118 or FOXO #1. Metabolite levels were compared after 14 days of feeding flies the HY + RU or HY - RU diet. (D) ROS production of hindgut (HG) cells was determined by DHE staining. After 14 days on the HY + RU or HY - RU diet entire guts from DaGS>_Uro_-RNAi #1 flies were isolated and stained with DHE ex vivo before the corrected total cell fluorescence (CTCF) of hindgut cells was determined according to reference [72]. (E) Dissected guts and Malpighian tubules (MT) from flies in (C) reared on the HY + RU diet for 14 days were stained ex vivo with DHE and imaged to visualize reactive oxygen intermediates. Image stitching was used to combine multiple confocal images with overlapping fields of view to produce a panorama view in high-resolution. The HG and the four tubes of the MT are indicated by a dashed line and white arrowheads, respectively. (F) DHE staining intensity of the MT and HG was quantified as in (D). Error bars represent the SE of multiple biological repeats.
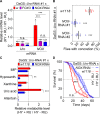
Fig 6. NOX mediated ROS production triggers lifespan attenuation and UA concretion formation on a high yeast diet.
(A) Levels of NOX mRNA upon modulation of FOXO. Recombinant DaGS::_Uro_-RNAi #1 flies were crossed to w1118 (w; no additional UAS-locus), or active UAS-transgenes triggering either over-expression of FOXO or inhibition of NOX (_NOX_-RNAi) and fed the HY + RU or HY - RU diet before relative mRNA levels of urate oxidase (Uro) and NADPH oxidase (NOX) were determined by qRT-PCR. (B) Concretion formation of recombinant DaGS::_Uro_-RNAi #1 flies crossed to w1118 or one of the two _NOX_-RNAi lines after 14 days of feeding the HY + RU diet. (C) Relative purine metabolite levels of fly homogenates from recombinant DaGS::_Uro_-RNAi #1 flies crossed to w1118 or _NOX_-RNAi #1. Metabolite levels were compared after 14 days of feeding flies the HY + RU or HY - RU diet. (D) Survival curves of flies from (C). Error bars represent the SE of multiple biological repeats. Supporting data is shown in S4 Fig and S5 Fig.
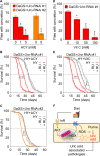
Fig 7. Pharmacological NOX inhibition phenocopies its genetic ablation and reduces UA related pathologies.
(A) Concretion formation of DaGS>_Uro_-RNAi #1 or DaGS>_Uro_-RNAi #2 flies after 14 days of feeding a HY + RU diet supplemented with the indicated concentration of the NOX inhibitor apocynin (ACY). (B, C) Survival curves of DaGS>_Uro_-RNAi #1 (B) or DaGS>_Uro_-RNAi #2 (C) flies fed the HY diet without or with 5 mM ACY (+ACY) supplementation in presence or absence of RU. (D) Concretion formation of DaGS>_Uro_-RNAi #1 flies after 14 days of feeding a HY + RU diet supplemented with the indicated concentration of the ROS scavenger vitamin C (Vit C). (E) As in (B), but 50 mM Vit C (+VitC) supplementation. (F) Key players influencing UA levels and associated pathologies are summarized. See text for more details. Error bars represent the SE of multiple biological repeats.
Similar articles
- A fly GWAS for purine metabolites identifies human FAM214 homolog medusa, which acts in a conserved manner to enhance hyperuricemia-driven pathologies by modulating purine metabolism and the inflammatory response.
Hilsabeck TAU, Liu-Bryan R, Guo T, Wilson KA, Bose N, Raftery D, Beck JN, Lang S, Jin K, Nelson CS, Oron T, Stoller M, Promislow D, Brem RB, Terkeltaub R, Kapahi P. Hilsabeck TAU, et al. Geroscience. 2022 Aug;44(4):2195-2211. doi: 10.1007/s11357-022-00557-9. Epub 2022 Apr 6. Geroscience. 2022. PMID: 35381951 Free PMC article. - Anthropometric variables, physical activity and dietary intakes of patients with uric acid nephrolithiasis.
Trinchieri A, Croppi E, Simonelli G, Sciorio C, Montanari E. Trinchieri A, et al. Urolithiasis. 2020 Apr;48(2):123-129. doi: 10.1007/s00240-019-01138-w. Epub 2019 Apr 29. Urolithiasis. 2020. PMID: 31037403 - Pathogenesis of gout.
Choi HK, Mount DB, Reginato AM; American College of Physicians; American Physiological Society. Choi HK, et al. Ann Intern Med. 2005 Oct 4;143(7):499-516. doi: 10.7326/0003-4819-143-7-200510040-00009. Ann Intern Med. 2005. PMID: 16204163 Review. No abstract available. - Uric acid and diet--insights into the epidemic of cardiovascular disease.
Johnson RJ, Rideout BA. Johnson RJ, et al. N Engl J Med. 2004 Mar 11;350(11):1071-3. doi: 10.1056/NEJMp048015. N Engl J Med. 2004. PMID: 15014177 No abstract available. - Implications of disorders of purine metabolism for the kidney and the urinary tract.
de Vries A, Sperling O. de Vries A, et al. Ciba Found Symp. 1977;(48):179-206. doi: 10.1002/9780470720301.ch12. Ciba Found Symp. 1977. PMID: 24529 Review.
Cited by
- Uric acid metabolism modulates diet-dependent responses to intraspecific competition in Drosophila larvae.
Morimoto J. Morimoto J. iScience. 2022 Nov 15;25(12):105598. doi: 10.1016/j.isci.2022.105598. eCollection 2022 Dec 22. iScience. 2022. PMID: 36458254 Free PMC article. - Circulating microRNA alternations in primary hyperuricemia and gout.
Bohatá J, Horváthová V, Pavlíková M, Stibůrková B. Bohatá J, et al. Arthritis Res Ther. 2021 Jul 10;23(1):186. doi: 10.1186/s13075-021-02569-w. Arthritis Res Ther. 2021. PMID: 34246297 Free PMC article. - Evaluating the beneficial effects of dietary restrictions: A framework for precision nutrigeroscience.
Wilson KA, Chamoli M, Hilsabeck TA, Pandey M, Bansal S, Chawla G, Kapahi P. Wilson KA, et al. Cell Metab. 2021 Nov 2;33(11):2142-2173. doi: 10.1016/j.cmet.2021.08.018. Epub 2021 Sep 22. Cell Metab. 2021. PMID: 34555343 Free PMC article. Review. - University of Southern California and buck institute nathan shock center: multidimensional models of aging.
Curran SP, Lithgow GJ, Verdin E, P C. Curran SP, et al. Geroscience. 2021 Oct;43(5):2119-2127. doi: 10.1007/s11357-021-00416-z. Epub 2021 Jul 16. Geroscience. 2021. PMID: 34269983 Free PMC article. - Genetic and metabolomic architecture of variation in diet restriction-mediated lifespan extension in Drosophila.
Jin K, Wilson KA, Beck JN, Nelson CS, Brownridge GW 3rd, Harrison BR, Djukovic D, Raftery D, Brem RB, Yu S, Drton M, Shojaie A, Kapahi P, Promislow D. Jin K, et al. PLoS Genet. 2020 Jul 9;16(7):e1008835. doi: 10.1371/journal.pgen.1008835. eCollection 2020 Jul. PLoS Genet. 2020. PMID: 32644988 Free PMC article.
References
- Mandal AK, Mount DB. The Molecular Physiology of Uric Acid Homeostasis. Annual Review of Physiology. 2015;77(1):323–45. - PubMed
- Kratzer JT, Lanaspa MA, Murphy MN, Cicerchi C, Graves CL, Tipton PA, et al. Evolutionary history and metabolic insights of ancient mammalian uricases. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(10):3763–8. Epub 2014/02/18. 10.1073/pnas.1320393111 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous