Translation of peptidoglycan metabolites into immunotherapeutics - PubMed (original) (raw)
Review
. 2019 Nov 29;8(12):e1095.
doi: 10.1002/cti2.1095. eCollection 2019.
Affiliations
- PMID: 31798878
- PMCID: PMC6883908
- DOI: 10.1002/cti2.1095
Review
Translation of peptidoglycan metabolites into immunotherapeutics
Matthew E Griffin et al. Clin Transl Immunology. 2019.
Abstract
The discovery of defined peptidoglycan metabolites that activate host immunity and their specific receptors has revealed fundamental insights into host-microbe recognition and afforded new opportunities for therapeutic development against infection and cancer. In this review, we summarise the discovery of two key peptidoglycan metabolites, γ-d-glutamyl-_meso_-diaminopimelic acid (iE-DAP) and muramyl dipeptide and their respective receptors, Nod1 and Nod2, and review progress towards translating these findings into therapeutic agents. Notably, synthetic derivatives of peptidoglycan metabolites have already yielded approved drugs for chemotherapy-induced leukopenia and paediatric osteosarcoma; however, the broad effects of peptidoglycan metabolites on host immunity suggest additional translational opportunities for new therapeutics towards other cancers, microbial infections and inflammatory diseases.
Keywords: adjuvant; cancer; infection; microbiota; pattern recognition receptor; peptidoglycan.
© 2019 The Authors. Clinical & Translational Immunology published by John Wiley & Sons Australia, Ltd on behalf of Australian and New Zealand Society for Immunology, Inc.
Conflict of interest statement
MEG and HCH are inventors on a patent filed by The Rockefeller University for the use of SagA towards the treatment of cancer and infection. Rise Therapeutics has licensed the patent on SagA for probiotic development.
Figures
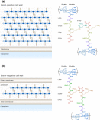
Figure 1
Structure of Gram‐positive and Gram‐negative peptidoglycan. (a) Gram‐positive bacteria like Enterococcus faecium contain a thick layer of peptidoglycan outside of their single membrane. Gram‐positive bacteria usually contain
d
‐isoglutamine (
d
‐iGln) and L‐lysine at the second and third positions of the peptide stem. (b) Gram‐negative bacteria such as Escherichia coli contain a thin layer of peptidoglycan within their periplasm. Gram‐negative bacteria generally utilise
d
‐glutamate (
d
‐Glu) and _meso_‐diaminopimelic acid (mDAP) at the second and third positions of the peptide stem. The location and composition of the crosslink between peptide stems vary between species. Ac, acetyl.
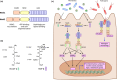
Figure 2
Peptidoglycan pattern recognition receptors. (a) Nucleotide‐binding oligomerisation domains 1 and 2 (Nod1 and Nod2) are conserved pattern recognition receptors of peptidoglycan fragments. Both proteins contain 1 or 2 caspase activation and recruitment domains (CARD), a NOD, and 10 or 11 leucine‐rich repeat (LRR) domains. (b) Nod1 recognises peptidoglycan fragments containing mDAP, the smallest of which is iE‐DAP 1. Conversely, Nod2 recognises muropeptides, with MDP 2 as the minimal active unit. (c) iE‐DAP and MDP derived from local bacteria or pathogens bind to Nod1 and Nod2, respectively. Activated Nod receptors oligomerise and recruit RIPK2. Through downstream adapter proteins and signalling cascades, Nod receptors activate NF‐κB, MAPK and IRF7 pathways to elicit expression of proinflammatory cytokines, antimicrobial peptides and type I interferon genes. Nod receptors also localise to sites of bacterial invasion to recruit ATG16L1 and induce autophagy pathways against the invading bacteria, known as xenophagy.
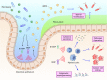
Figure 3
Peptidoglycan activation leads to diverse immune responses**.** Peptidoglycan fragments iE‐DAP and MDP exhibit a wide array of activities through the activation of numerous cell types. Activation of intestinal epithelial cells leads to the increased production of mucins and antimicrobial peptides (AMPs) to improve the intestinal epithelial barrier and prevent infection. Activation of different myeloid cell populations can lead to increased cytokine secretion, epigenetic reprogramming, and the direct killing of pathogens and other foreign‐presenting cells such as cancer cells.
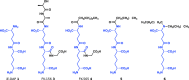
Figure 4
Representative iE‐DAP‐derived Nod1 agonists. The chemical structures of iE‐DAP and representative Nod1 agonists are shown. In all structures, the iE‐DAP scaffold is highlighted in blue. In general, a trend towards higher lipophilicity has resulted in increased biological activity.
Figure 5
Representative MDP‐derived Nod2 agonists. The chemical structures of MDP and representative Nod2 agonists are shown. In all structures, the MDP scaffold is highlighted in green. As seen with Nod1‐targeted molecules, an increase in lipophilicity has resulted in more potent agonists. Modifications such as esterification also have decreased side effects such as pyrogenicity observed with the parental MDP molecule. Moreover, departure from the MurNAc monosaccharide has yielded compounds with similar efficacy. _n_‐Bu, _n_‐butyl; Et, ethyl.
Similar articles
- Peptidoglycan Metabolite Photoaffinity Reporters Reveal Direct Binding to Intracellular Pattern Recognition Receptors and Arf GTPases.
Wang YC, Westcott NP, Griffin ME, Hang HC. Wang YC, et al. ACS Chem Biol. 2019 Mar 15;14(3):405-414. doi: 10.1021/acschembio.8b01038. Epub 2019 Feb 20. ACS Chem Biol. 2019. PMID: 30735346 Free PMC article. - The frameshift mutation in Nod2 results in unresponsiveness not only to Nod2- but also Nod1-activating peptidoglycan agonists.
Netea MG, Ferwerda G, de Jong DJ, Werts C, Boneca IG, Jéhanno M, Van Der Meer JW, Mengin-Lecreulx D, Sansonetti PJ, Philpott DJ, Dharancy S, Girardin SE. Netea MG, et al. J Biol Chem. 2005 Oct 28;280(43):35859-67. doi: 10.1074/jbc.M504924200. Epub 2005 Aug 22. J Biol Chem. 2005. PMID: 16115863 - An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid.
Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nuñez G, Inohara N. Chamaillard M, et al. Nat Immunol. 2003 Jul;4(7):702-7. doi: 10.1038/ni945. Epub 2003 Jun 6. Nat Immunol. 2003. PMID: 12796777 - Chemical synthesis of peptidoglycan fragments for elucidation of the immunostimulating mechanism.
Fujimoto Y, Inamura S, Kawasaki A, Shiokawa Z, Shimoyama A, Hashimoto T, Kusumoto S, Fukase K. Fujimoto Y, et al. J Endotoxin Res. 2007;13(3):189-96. doi: 10.1177/0968051907080739. J Endotoxin Res. 2007. PMID: 17621561 Review. - Intracellular NOD-like receptors in host defense and disease.
Kanneganti TD, Lamkanfi M, Núñez G. Kanneganti TD, et al. Immunity. 2007 Oct;27(4):549-59. doi: 10.1016/j.immuni.2007.10.002. Immunity. 2007. PMID: 17967410 Review.
Cited by
- Structural Fine-Tuning of Desmuramylpeptide NOD2 Agonists Defines Their In Vivo Adjuvant Activity.
Guzelj S, Nabergoj S, Gobec M, Pajk S, Klančič V, Slütter B, Frkanec R, Štimac A, Šket P, Plavec J, Mlinarič-Raščan I, Jakopin Ž. Guzelj S, et al. J Med Chem. 2021 Jun 10;64(11):7809-7838. doi: 10.1021/acs.jmedchem.1c00644. Epub 2021 May 27. J Med Chem. 2021. PMID: 34043358 Free PMC article. - Enterococcus NlpC/p60 Peptidoglycan Hydrolase SagA Localizes to Sites of Cell Division and Requires Only a Catalytic Dyad for Protease Activity.
Espinosa J, Lin TY, Estrella Y, Kim B, Molina H, Hang HC. Espinosa J, et al. Biochemistry. 2020 Nov 24;59(46):4470-4480. doi: 10.1021/acs.biochem.0c00755. Epub 2020 Nov 2. Biochemistry. 2020. PMID: 33136372 Free PMC article. - Distinctive Immune Signatures Driven by Structural Alterations in Desmuramylpeptide NOD2 Agonists.
Janež Š, Guzelj S, Kocbek P, de Vlieger EA, Slütter B, Jakopin Ž. Janež Š, et al. J Med Chem. 2024 Oct 10;67(19):17585-17607. doi: 10.1021/acs.jmedchem.4c01577. Epub 2024 Sep 29. J Med Chem. 2024. PMID: 39344184 Free PMC article. - The amalgamation of cellular metabolism and immunology for host immunity.
Mintern JD, Binger KJ. Mintern JD, et al. Clin Transl Immunology. 2020 Mar 11;9(3):e1123. doi: 10.1002/cti2.1123. eCollection 2020. Clin Transl Immunology. 2020. PMID: 32190325 Free PMC article. - Innate Immune Response against Hepatitis C Virus: Targets for Vaccine Adjuvants.
Sepulveda-Crespo D, Resino S, Martinez I. Sepulveda-Crespo D, et al. Vaccines (Basel). 2020 Jun 17;8(2):313. doi: 10.3390/vaccines8020313. Vaccines (Basel). 2020. PMID: 32560440 Free PMC article. Review.
References
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature 2007; 449: 819–826. - PubMed
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140: 805–820. - PubMed
- Wolf AJ, Underhill DM. Peptidoglycan recognition by the innate immune system. Nat Rev Immunol 2018; 18: 243–254. - PubMed
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev 2008; 32: 149–167. - PubMed
Publication types
Grants and funding
- F32 AT010087/AT/NCCIH NIH HHS/United States
- R01 CA245292/CA/NCI NIH HHS/United States
- R01 GM103593/GM/NIGMS NIH HHS/United States
- R21 AI156674/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources