Insights into Drought Stress Signaling in Plants and the Molecular Genetic Basis of Cotton Drought Tolerance - PubMed (original) (raw)
Review
Insights into Drought Stress Signaling in Plants and the Molecular Genetic Basis of Cotton Drought Tolerance
Tahir Mahmood et al. Cells. 2019.
Abstract
Drought stress restricts plant growth and development by altering metabolic activity and biological functions. However, plants have evolved several cellular and molecular mechanisms to overcome drought stress. Drought tolerance is a multiplex trait involving the activation of signaling mechanisms and differentially expressed molecular responses. Broadly, drought tolerance comprises two steps: stress sensing/signaling and activation of various parallel stress responses (including physiological, molecular, and biochemical mechanisms) in plants. At the cellular level, drought induces oxidative stress by overproduction of reactive oxygen species (ROS), ultimately causing the cell membrane to rupture and stimulating various stress signaling pathways (ROS, mitogen-activated-protein-kinase, Ca2+, and hormone-mediated signaling). Drought-induced transcription factors activation and abscisic acid concentration co-ordinate the stress signaling and responses in cotton. The key responses against drought stress, are root development, stomatal closure, photosynthesis, hormone production, and ROS scavenging. The genetic basis, quantitative trait loci and genes of cotton drought tolerance are presented as examples of genetic resources in plants. Sustainable genetic improvements could be achieved through functional genomic approaches and genome modification techniques such as the CRISPR/Cas9 system aid the characterization of genes, sorted out from stress-related candidate single nucleotide polymorphisms, quantitative trait loci, and genes. Exploration of the genetic basis for superior candidate genes linked to stress physiology can be facilitated by integrated functional genomic approaches. We propose a third-generation sequencing approach coupled with genome-wide studies and functional genomic tools, including a comparative sequenced data (transcriptomics, proteomics, and epigenomic) analysis, which offer a platform to identify and characterize novel genes. This will provide information for better understanding the complex stress cellular biology of plants.
Keywords: Gossypium; cellular stress signaling; cotton molecular genetic basis; drought stress responses; drought tolerance; functional genomics; gene identification tools.
Conflict of interest statement
The authors declare that there is no conflict of interests regarding the publication of this manuscript.
Figures
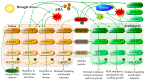
Figure 1
Drought-induced cellular and molecular signaling pathways to enhance drought tolerance in plants. The cell membrane perceives stress signals and triggers signaling. In the presence of abscisic acid (ABA), a complex of PRY/PRL, RCARs, and PP2Cs is formed, which dissociates PP2Cs from SnRK2 and activates NnRK2 protein (P). SnRK2 is auto-activated when separated from PP2C. Activated SnRK2 triggers and regulates molecular and physiological responses. Similarly, jasmonic acid (JA) is engaged with the jasmonate-zim domain (JAZ) in a complex with SCF and TFs (MYC2), and activates stress- responsive genes. Overproduction of reactive oxygen species (ROS) in response to oxo-phytodienoic acid (OPDA) and JA activates scavenging genes and act like a stress-signaling unit. Calcium (Ca2+) interacts with mitogen-activated protein kinase (MAPK) cascade proteins to activate transcriptional factors and signaling genes. Finally, functional proteins (FP) are synthesized for drought-stress responses.
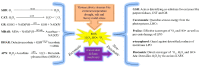
Figure 2
Drought-induced, ABA-dependent, ABA-independent MAPK signaling, and interaction between ABA, ROS, and MAPK signaling under drought stress in plants. ABA-regulates various MAPKs in cotton and Arabidopsis. ABA promotes drought sensing and signaling in plants. The different cascades are represented by different color schemes in the figure. Solid arrow lines denote established signaling mechanisms, while dashed arrow lines denote unestablished signaling pathways. ABA-activated SnRK2s (See Figure 1 for SnRK2 activation) trigger and phosphorylate downstream targets, such as respiratory burst oxidase homolog (RBOH) and various MAPKs. Activation of RBOH induces ROS production. ROS signaling and ABA signaling may overlap with MAPK factors, to interact and regulate drought tolerance. MAP3K17/18-MKK3-MPK1/2/7/14 is an ABA-induced complete MAPK cascade involved in stomatal signaling, senescence, and drought tolerance mechanisms in Arabidopsis. In addition, MKK1 activates MPK6 to positively regulate CATALASE1 (CAT1) for ROS abundance. In cotton, the drought- and ABA-induced MAPK cascade MKK3-MPK7-PIP1 is associated with stomatal signaling and drought tolerance. Another ABA-mediated MAPK module, MAPKKK49-MKK4/MKK5, is associated with abiotic stress responses. GhMPK17 gene is a novel, well-characterized MAPK, which is associated with responses to osmotic and salt stresses in cotton. An ABA-independent and drought-mediated MAPK module (MAP3K15-MKK4-MPK6-WRKY59) regulates drought tolerance in cotton. Drought stress triggers the MAPKKK15 cascade, which phosphorylates the WRKY59 transcriptional factor. Interestingly, WRKY59 binds to the promoter of DREB2 and regulates the expression of drought-sensitive genes. Meanwhile, it positively regulates the expression of MAP3K15 by establishing a feedback loop, which regulates drought tolerance in cotton.
Figure 3
Overall pathways of drought stress effects and plant responses to drought stress.
Figure 4
Anti-oxidant machinery scavenges cellular reactive oxygen species (ROS) through two pathways in plants. One is the enzymatic pathway and the other is a non-enzymatic pathway. Several enzymes convert ROS to non-harmful substances via enzymatic pathways in plant cells, while in the non-enzymatic pathway, other substances convert ROS to non-harmful substances.
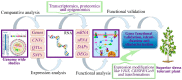
Figure 5
Schematic flow of research processes from genome-wide studies to functional validation, characterization, and identification of drought stress-responsive genes in plants. QTL, quantitative trait loci, CNV, copy number variation, SNP, single nucleotide polymorphism, mRNA, messenger RNA, SRNA, small RNA, DAP, differential abundant proteins, and DEG, differential expressed genes.
Similar articles
- Drought coping strategies in cotton: increased crop per drop.
Ullah A, Sun H, Yang X, Zhang X. Ullah A, et al. Plant Biotechnol J. 2017 Mar;15(3):271-284. doi: 10.1111/pbi.12688. Plant Biotechnol J. 2017. PMID: 28055133 Free PMC article. Review. - Comparative Transcriptomic Analysis of Biological Process and Key Pathway in Three Cotton (Gossypium spp.) Species Under Drought Stress.
Hasan MM, Ma F, Islam F, Sajid M, Prodhan ZH, Li F, Shen H, Chen Y, Wang X. Hasan MM, et al. Int J Mol Sci. 2019 Apr 27;20(9):2076. doi: 10.3390/ijms20092076. Int J Mol Sci. 2019. PMID: 31035558 Free PMC article. - Genome-wide functional analysis of cotton (Gossypium hirsutum) in response to drought.
Chen Y, Liu ZH, Feng L, Zheng Y, Li DD, Li XB. Chen Y, et al. PLoS One. 2013 Nov 18;8(11):e80879. doi: 10.1371/journal.pone.0080879. eCollection 2013. PLoS One. 2013. PMID: 24260499 Free PMC article. - Mechanism of Stomatal Closure in Plants Exposed to Drought and Cold Stress.
Agurla S, Gahir S, Munemasa S, Murata Y, Raghavendra AS. Agurla S, et al. Adv Exp Med Biol. 2018;1081:215-232. doi: 10.1007/978-981-13-1244-1_12. Adv Exp Med Biol. 2018. PMID: 30288712 Review.
Cited by
- The BBX gene CmBBX22 negatively regulates drought stress tolerance in chrysanthemum.
Liu Y, Cheng H, Cheng P, Wang C, Li J, Liu Y, Song A, Chen S, Chen F, Wang L, Jiang J. Liu Y, et al. Hortic Res. 2022 Aug 25;9:uhac181. doi: 10.1093/hr/uhac181. eCollection 2022. Hortic Res. 2022. PMID: 36338842 Free PMC article. - Calcium-Dependent Protein Kinase GhCDPK16 Exerts a Positive Regulatory Role in Enhancing Drought Tolerance in Cotton.
Yan M, Chai M, Li L, Dong Z, Jin H, Tan M, Ye Z, Yu S, Feng Z. Yan M, et al. Int J Mol Sci. 2024 Jul 30;25(15):8308. doi: 10.3390/ijms25158308. Int J Mol Sci. 2024. PMID: 39125876 Free PMC article. - ERF subfamily transcription factors and their function in plant responses to abiotic stresses.
Wu Y, Li X, Zhang J, Zhao H, Tan S, Xu W, Pan J, Yang F, Pi E. Wu Y, et al. Front Plant Sci. 2022 Nov 30;13:1042084. doi: 10.3389/fpls.2022.1042084. eCollection 2022. Front Plant Sci. 2022. PMID: 36531407 Free PMC article. Review. - Crop Proteomics under Abiotic Stress: From Data to Insights.
Kausar R, Wang X, Komatsu S. Kausar R, et al. Plants (Basel). 2022 Oct 27;11(21):2877. doi: 10.3390/plants11212877. Plants (Basel). 2022. PMID: 36365330 Free PMC article. Review.
References
- Saranga Y., Paterson A.H., Levi A. Bridging Classical and Molecular Genetics of Abiotic Stress Resistance in Cotton. Genet. Genom. Cott. 2009;3:337–352.
- Abdelraheem A., Esmaeili N., O’Connell M., Zhang J. Progress and perspective on drought and salt stress tolerance in cotton. Ind. Crops Prod. 2019;130:118–129. doi: 10.1016/j.indcrop.2018.12.070. - DOI
- Li L., Yu D., Zhao F., Pang C., Song M., Wei H., Fan S., Yu S. Genome-wide analysis of the calcium-dependent protein kinase gene family in Gossypium raimondii. J. Integr. Agric. 2015;14:29–41. doi: 10.1016/S2095-3119(14)60780-2. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous