LFA-1 signals to promote actin polymerization and upstream migration in T cells - PubMed (original) (raw)
LFA-1 signals to promote actin polymerization and upstream migration in T cells
Nathan H Roy et al. J Cell Sci. 2020.
Abstract
T cell entry into inflamed tissue requires firm adhesion, cell spreading, and migration along and through the endothelial wall. These events require the T cell integrins LFA-1 and VLA-4 and their endothelial ligands ICAM-1 and VCAM-1, respectively. T cells migrate against the direction of shear flow on ICAM-1 and with the direction of shear flow on VCAM-1, suggesting that these two ligands trigger distinct cellular responses. However, the contribution of specific signaling events downstream of LFA-1 and VLA-4 has not been explored. Using primary mouse T cells, we found that engagement of LFA-1, but not VLA-4, induces cell shape changes associated with rapid 2D migration. Moreover, LFA-1 ligation results in activation of the phosphoinositide 3-kinase (PI3K) and ERK pathways, and phosphorylation of multiple kinases and adaptor proteins, whereas VLA-4 ligation triggers only a subset of these signaling events. Importantly, T cells lacking Crk adaptor proteins, key LFA-1 signaling intermediates, or the ubiquitin ligase cCbl (also known as CBL), failed to migrate against the direction of shear flow on ICAM-1. These studies identify novel signaling differences downstream of LFA-1 and VLA-4 that drive T cell migratory behavior.This article has an associated First Person interview with the first author of the paper.
Keywords: Actin; Integrins; Migration; Shear flow; Signaling; T cell.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
Fig. 1.
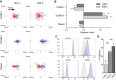
Activated primary mouse T cells migrate against the direction of shear on ICAM-1. (A) Representative scattergrams of activated primary mouse CD4+ T cells migrating on ICAM-1, VCAM-1, or mixed surfaces under shear flow conditions (100 s−1 or 800 s−1). Red lines, cell tracks with a net migration against the direction of shear (upstream). Blue lines, cell tracks with a net migration in the direction of shear (downstream). (B) Average migration index (MI) pooled from three independent experiments. MI for each cell equals the ratio of the displacement left or right by the total distance traveled. Migration with no directional preference results in a value of 0. Negative values indicate net migration upstream, and positive values indicate downstream migration. (C) Surface levels of CD11a and CD18 (integrins αL and β2, respectively, which compose LFA-1), and CD49d and CD29 (integrins α4 and β1, respectively, which compose VLA-4) on CD4+ T cells at day 5 post activation (the day used for all migration experiments). Gray shaded lines represent isotype controls. (D) CD4+ T cell adhesion to the indicated ligands was measured using a plate-based adhesion assay. B and D show mean±s.d. for three independent experiments. Statistics were calculated using a one-way ANOVA with a Tukey multiple comparisons test. **P<0.01; ***P<0.001.
Fig. 2.
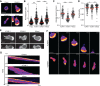
Quantitative and qualitative differences in T cell migration on ICAM-1 and VCAM-1. (A) CD4+ T cells were allowed to migrate on the indicated ligands under static conditions, then were fixed and stained with phalloidin to visualize F-actin. Images are a projection of four _z_-slices totaling 0.75 μm, displayed as heat maps based on pixel intensity (high intensity, yellow; low intensity, violet). PolyL, poly-L-lysine control. Scale bar: 10 μm. (B) Quantification of total F-actin intensity per cell (each circle represents one cell). (C,D) T cells were tracked while migrating on the indicated surfaces in the absence of shear, and the (C) average speed and (D) directionality were calculated (each circle represents one cell). (E) Orthogonal views of T cells expressing Lifeact–GFP, migrating on surfaces coated with the indicated ligands. Scale bar: 10 μm. (F) Migrating T cells expressing Lifeact–GFP were imaged using time-lapse confocal microscopy. Images are a projection of a 2 μm total stack starting at the cell–coverslip interface, and are displayed as heat maps. Scale bar: 10 μm. See
Movies 1
and
2
. (G) Kymographs of T cells migrating as in F. Data in B–D are pooled from three independent experiments, and red lines indicate the mean. Statistics were calculated using a one-way ANOVA with a Tukey multiple comparisons test. *P<0.05; **P<0.01; ***P<0.001.
Fig. 3.
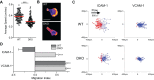
Crk-deficient T cells fail to migrate upstream on ICAM-1. (A,B) WT and DKO T cells were imaged while migrating on ICAM-1 in the absence of shear. (A) Average speed pooled from three independent experiments (each circle represents one cell, and red lines indicate the mean). (B) Representative images of migrating T cells expressing Lifeact–GFP, displayed as in Fig. 2. Scale bar: 10 μm. (C) Representative scattergrams of WT and DKO T cells imaged while migrating on ICAM-1 or VCAM-1 under shear flow (shear rate 800 s−1). Red lines, cell tracks with a net migration against the direction of shear (upstream). Blue lines, cell tracks with a net migration in the direction of shear (downstream). (D) Average migration index calculated from three independent experiments, displayed as mean±s.d. Statistics were calculated using a one-way ANOVA with a Tukey multiple comparisons test. ***P<0.001.
Fig. 4.
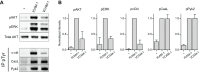
Differential signaling induced by ICAM-1 and VCAM-1. (A) CD4+ T cells were allowed to settle on ICAM-1- or VCAM-1-coated plates for 20 min and lysed. Lysates were immunoblotted with the indicated antibodies (top panel), or immunoprecipitated with anti-pTyr, and immunoblotted with indicated antibodies (bottom panel). −, lysates from unstimulated cells. (B) Quantification of immunoblots performed as in A. Data represent mean±s.d., normalized to the ICAM-1 condition from three independent experiments.
Fig. 5.
cCbl knockout reverses upstream migration on ICAM-1. (A) CD4+ T cells from Cas9-expressing mice were transduced with the indicated gRNAs, selected for 3 d in puromycin, and then lysed and immunoblotted for the indicated proteins. NT, non-targeting gRNA control. GAPDH was used as a loading control. (B) Quantification of immunoblots performed as in A. Data represent mean±s.d., normalized to the NT control, from three independent experiments. (C,D) T cells expressing the indicated gRNAs were allowed to migrate on ICAM-1-coated surfaces, fixed, and stained with fluorescent phalloidin. (C) Representative images, displayed as in Fig. 2. Scale bar: 10 μm. (D) Quantification of F-actin intensity per cell, pooled from three independent experiments (each circle represents one cell, and red lines indicate the mean). (E) Migration index of T cells expressing the indicated gRNAs migrating on ICAM-1 under shear flow (shear rate 800 s−1). (F) Percentage of cells migrating upstream from experiments shown in panel E. (G) Distribution of cells with a given MI pooled from three independent experiments. E and F show mean±s.e.m. for three independent experiments. Statistics were calculated using a one-way ANOVA with a Tukey multiple comparisons test. ns, not significant; *P<0.05; **P<0.01; ***P<0.001.
Similar articles
- Integrin crosstalk allows CD4+ T lymphocytes to continue migrating in the upstream direction after flow.
Kim SHJ, Hammer DA. Kim SHJ, et al. Integr Biol (Camb). 2019 Dec 31;11(10):384-393. doi: 10.1093/intbio/zyz034. Integr Biol (Camb). 2019. PMID: 31851360 Free PMC article. - Crk adaptor proteins mediate actin-dependent T cell migration and mechanosensing induced by the integrin LFA-1.
Roy NH, MacKay JL, Robertson TF, Hammer DA, Burkhardt JK. Roy NH, et al. Sci Signal. 2018 Dec 11;11(560):eaat3178. doi: 10.1126/scisignal.aat3178. Sci Signal. 2018. PMID: 30538176 Free PMC article. - Lymphocyte adhesion and transendothelial migration in the central nervous system: the role of LFA-1, ICAM-1, VLA-4 and VCAM-1. off.
Greenwood J, Wang Y, Calder VL. Greenwood J, et al. Immunology. 1995 Nov;86(3):408-15. Immunology. 1995. PMID: 8550078 Free PMC article. - Inhibition of LFA-1/ICAM-1 and VLA-4/VCAM-1 as a therapeutic approach to inflammation and autoimmune diseases.
Yusuf-Makagiansar H, Anderson ME, Yakovleva TV, Murray JS, Siahaan TJ. Yusuf-Makagiansar H, et al. Med Res Rev. 2002 Mar;22(2):146-67. doi: 10.1002/med.10001. Med Res Rev. 2002. PMID: 11857637 Review.
Cited by
- Inhibition of Mac-1 allows human macrophages to migrate against the direction of shear flow on ICAM-1.
Mittal A, Guin S, Mochida A, Hammer DA, Buffone A Jr. Mittal A, et al. Mol Biol Cell. 2024 Oct 1;35(10):br18. doi: 10.1091/mbc.E24-03-0114. Epub 2024 Aug 21. Mol Biol Cell. 2024. PMID: 39167496 - CellTracksColab is a platform that enables compilation, analysis, and exploration of cell tracking data.
Gómez-de-Mariscal E, Grobe H, Pylvänäinen JW, Xénard L, Henriques R, Tinevez JY, Jacquemet G. Gómez-de-Mariscal E, et al. PLoS Biol. 2024 Aug 8;22(8):e3002740. doi: 10.1371/journal.pbio.3002740. eCollection 2024 Aug. PLoS Biol. 2024. PMID: 39116189 Free PMC article. - Mechanical regulation of lymphocyte activation and function.
Pathni A, Wagh K, Rey-Suarez I, Upadhyaya A. Pathni A, et al. J Cell Sci. 2024 Jul 1;137(13):jcs219030. doi: 10.1242/jcs.219030. Epub 2024 Jul 12. J Cell Sci. 2024. PMID: 38995113 Review. - Tumor-intrinsic IFNα and CXCL10 are critical for immunotherapeutic efficacy by recruiting and activating T lymphocytes in tumor microenvironment.
Cheng CC, Chang J, Ho AS, Sie ZL, Peng CL, Wang CL, Dev K, Chang CC. Cheng CC, et al. Cancer Immunol Immunother. 2024 Jul 2;73(9):175. doi: 10.1007/s00262-024-03761-y. Cancer Immunol Immunother. 2024. PMID: 38953994 Free PMC article. - cellPLATO - an unsupervised method for identifying cell behaviour in heterogeneous cell trajectory data.
Shannon MJ, Eisman SE, Lowe AR, Sloan TFW, Mace EM. Shannon MJ, et al. J Cell Sci. 2024 Oct 15;137(20):jcs261887. doi: 10.1242/jcs.261887. Epub 2024 Jun 12. J Cell Sci. 2024. PMID: 38738282 Free PMC article.
References
- Bartholomäus I., Kawakami N., Odoardi F., Schlager C., Miljkovic D., Ellwart J. W., Klinkert W. E., Flugel-Koch C., Issekutz T. B., Wekerle H. et al. (2009). Effector T cell interactions with meningeal vascular structures in nascent autoimmune CNS lesions. Nature 462, 94-98. 10.1038/nature08478 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA009140/CA/NCI NIH HHS/United States
- T32 AR007442/AR/NIAMS NIH HHS/United States
- R01 GM104867/GM/NIGMS NIH HHS/United States
- R01 HL128551/HL/NHLBI NIH HHS/United States
- R01 GM123019/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous