Population structure, biogeography and transmissibility of Mycobacterium tuberculosis - PubMed (original) (raw)
. 2021 Oct 20;12(1):6099.
doi: 10.1038/s41467-021-26248-1.
Roger Vargas Jr 2 3, Ashaque Husain 4, S M Mostofa Kamal 5, Alena Skrahina 6, Sabira Tahseen 7, Nazir Ismail 8 9, Anna Barbova 10, Stefan Niemann 11, Daniela Maria Cirillo 12, Anna S Dean 13, Matteo Zignol 13, Maha Reda Farhat 14 15
Affiliations
- PMID: 34671035
- PMCID: PMC8528816
- DOI: 10.1038/s41467-021-26248-1
Population structure, biogeography and transmissibility of Mycobacterium tuberculosis
Luca Freschi et al. Nat Commun. 2021.
Abstract
Mycobacterium tuberculosis is a clonal pathogen proposed to have co-evolved with its human host for millennia, yet our understanding of its genomic diversity and biogeography remains incomplete. Here we use a combination of phylogenetics and dimensionality reduction to reevaluate the population structure of M. tuberculosis, providing an in-depth analysis of the ancient Indo-Oceanic Lineage 1 and the modern Central Asian Lineage 3, and expanding our understanding of Lineages 2 and 4. We assess sub-lineages using genomic sequences from 4939 pan-susceptible strains, and find 30 new genetically distinct clades that we validate in a dataset of 4645 independent isolates. We find a consistent geographically restricted or unrestricted pattern for 20 groups, including three groups of Lineage 1. The distribution of terminal branch lengths across the M. tuberculosis phylogeny supports the hypothesis of a higher transmissibility of Lineages 2 and 4, in comparison with Lineages 3 and 1, on a global scale. We define an expanded barcode of 95 single nucleotide substitutions that allows rapid identification of 69 M. tuberculosis sub-lineages and 26 additional internal groups. Our results paint a higher resolution picture of the M. tuberculosis phylogeny and biogeography.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
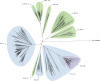
Fig. 1. Phylogenetic tree reconstruction of lineage 1 (binary tree).
Gray circles define splits where the _F_ST (fixation index) calculated using the descendants of the two children nodes is greater than 0.33. The sub-lineages are defined by colored areas (blue: sub-lineages already described in the literature; green: sub-lineages described here; purple: internal sub-lineages). Source data are provided as a Source Data file.
Fig. 2. Phylogenetic tree reconstruction of lineage 3 (binary tree).
Gray circles define splits where the _F_ST (fixation index) calculated using the descendants of the two children nodes is greater than 0.33. The sub-lineages are defined by colored areas (green: sub-lineages described here; purple: internal sub-lineages). Source data are provided as a Source Data file.
Fig. 3. Phylogenetic tree reconstruction of lineage 2 (binary tree).
Gray circles define splits where the _F_ST (fixation index) calculated using the descendants of the two children nodes is greater than 0.33. The sub-lineages are defined by colored areas (blue: sub-lineages already described in the literature; green: sub-lineages described here; purple: internal sub-lineages). Source data are provided as a Source Data file.
Fig. 4. Phylogenetic tree reconstruction of lineage 4 (binary tree).
Gray circles define splits where the _F_ST (fixation index) calculated using the descendants of the two children nodes is greater than 0.33. The sub-lineages are defined by colored areas (blue: sub-lineages already described in the literature; green: sub-lineages described here; purple: internal sub-lineages). Source data are provided as a Source Data file.
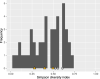
Fig. 5. Histogram of the Simpson diversity index calculated for sub-lineages of lineages 1–4.
A data set of 17,432 isolates from 74 countries was used to perform this analysis. Yellow triangles designate the Simpson diversity index values of sub-lineages designated as geographically restricted by Stucki et al. Light gray circles designate the Simpson diversity index values of sub-lineages designated as geographically unrestricted by Stucki et al. Source data are provided as a Source Data file.
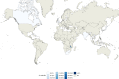
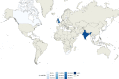
Fig. 6. Geographic distribution of internal sub-lineage 1.1.3.i1.
Colors represent the percentage of 1.1.3.i1 strains isolated in a given country with respect to all lineage 1 strains isolated in such country. Source data are provided as a Source Data file.
Fig. 7. Geographic distribution of internal sub-lineage 1.1.1.1.
Colors represent the percentage of 1.1.1.1 strains isolated in a given country with respect to all lineage 1 strains isolated in such country. Source data are provided as a Source Data file.
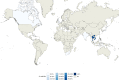
Fig. 8. Geographic distribution of internal sub-lineage 1.1.2.
Colors represent the percentage of 1.1.2 strains isolated in a given country with respect to all lineage 1 strains isolated in such country. Source data are provided as a Source Data file.
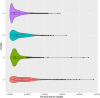
Fig. 9. Distributions of terminal branch lengths for the four global Mtb lineages (L1–L4).
Two-sided Wilcoxon rank sum tests were performed to test that two distributions were significantly different. Medians: 6.2 × 10−5 (L4), 8.2 × 10−5 (L2), 10.2 × 10−5 (L3), 17.5 × 10−5 (L1). Comparisons: L1 vs L2, L3 or L4 (_p_-value < 2.2 × 10−16); L2 vs L3 (_p_-value = 3.6 × 10−6), L2 vs L4 (_p_-value < 2.2 × 10−16); L3 vs L4 (_p_-value < 2.2 × 10−16). Description of the distributions (L1: n = 739, Min: 0.5 × 10−5, 1st Quartile: 6.7 × 10−5, Median: 17.5 × 10−5, 3rd Quartile: 28 × 10−5, Max: 120 × 10−5; L2: n = 2193, Min: 0.7 × 10−5, 1st Quartile: 5.3 × 10−5, Median: 8.2 × 10−5, 3rd Quartile: 12 × 10−5, Max: 110 × 10−5; L3: n = 1103, Min: 0.5 × 10−5, 1st Quartile: 4.5 × 10−5, Median: 10.2 × 10−5, 3rd Quartile: 20 × 10−5, Max: 80 × 10−5; L4: n = 5514, Min: 0.2 × 10−5, 1st Quartile: 2.6 × 10−5, Median: 6.2 × 10−5, 3rd Quartile: 13 × 10−5, Max: 70 × 10−5). Source data are provided as a Source Data file.
Similar articles
- Robust barcoding and identification of Mycobacterium tuberculosis lineages for epidemiological and clinical studies.
Napier G, Campino S, Merid Y, Abebe M, Woldeamanuel Y, Aseffa A, Hibberd ML, Phelan J, Clark TG. Napier G, et al. Genome Med. 2020 Dec 14;12(1):114. doi: 10.1186/s13073-020-00817-3. Genome Med. 2020. PMID: 33317631 Free PMC article. - [Population structure analysis of Mycobacterium tuberculosis Beijing family in Japan].
Iwamoto T. Iwamoto T. Kekkaku. 2009 Dec;84(12):755-9. Kekkaku. 2009. PMID: 20077859 Japanese. - A robust SNP barcode for typing Mycobacterium tuberculosis complex strains.
Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, Portugal I, Pain A, Martin N, Clark TG. Coll F, et al. Nat Commun. 2014 Sep 1;5:4812. doi: 10.1038/ncomms5812. Nat Commun. 2014. PMID: 25176035 Free PMC article. - Consequences of genomic diversity in Mycobacterium tuberculosis.
Coscolla M, Gagneux S. Coscolla M, et al. Semin Immunol. 2014 Dec;26(6):431-44. doi: 10.1016/j.smim.2014.09.012. Epub 2014 Oct 22. Semin Immunol. 2014. PMID: 25453224 Free PMC article. Review. - The Nature and Evolution of Genomic Diversity in the Mycobacterium tuberculosis Complex.
Brites D, Gagneux S. Brites D, et al. Adv Exp Med Biol. 2017;1019:1-26. doi: 10.1007/978-3-319-64371-7_1. Adv Exp Med Biol. 2017. PMID: 29116627 Review.
Cited by
- Discordance Between Phenotypic and WGS-Based Drug Susceptibility Testing Results for Some Anti-Tuberculosis Drugs: A Snapshot Study of Paired Mycobacterium tuberculosis Isolates with Small Genetic Distance.
Sadovska D, Nodieva A, Pole I, Vīksna A, Ķimsis J, Ozere I, Norvaiša I, Bogdanova I, Bandere D, Ranka R. Sadovska D, et al. Infect Drug Resist. 2024 Aug 1;17:3289-3307. doi: 10.2147/IDR.S468997. eCollection 2024. Infect Drug Resist. 2024. PMID: 39108991 Free PMC article. - Bedaquiline Resistance and Molecular Characterization of Rifampicin-Resistant Mycobacterium Tuberculosis Isolates in Zhejiang, China.
Tong E, Zhou Y, Liu Z, Zhu Y, Zhang M, Wu K, Pan J, Jiang J. Tong E, et al. Infect Drug Resist. 2023 Oct 31;16:6951-6963. doi: 10.2147/IDR.S429003. eCollection 2023. Infect Drug Resist. 2023. PMID: 37928607 Free PMC article. - Global-scale GWAS associates a subset of SNPs with animal-adapted variants in M. tuberculosis complex.
Brenner EP, Sreevatsan S. Brenner EP, et al. BMC Med Genomics. 2023 Oct 24;16(1):260. doi: 10.1186/s12920-023-01695-5. BMC Med Genomics. 2023. PMID: 37875894 Free PMC article. - Connection between two historical tuberculosis outbreak sites in Japan, Honshu, by a new ancestral Mycobacterium tuberculosis L2 sublineage.
Guyeux C, Senelle G, Refrégier G, Bretelle-Establet F, Cambau E, Sola C. Guyeux C, et al. Epidemiol Infect. 2022 Jan 19;150:1-25. doi: 10.1017/S0950268822000048. Online ahead of print. Epidemiol Infect. 2022. PMID: 35042579 Free PMC article. - Transmission and Drug Resistance Genotype of Multidrug-Resistant or Rifampicin-Resistant Mycobacterium tuberculosis in Chongqing, China.
Zhao B, Liu C, Fan J, Ma A, He W, Hu Y, Zhao Y. Zhao B, et al. Microbiol Spectr. 2022 Oct 26;10(5):e0240521. doi: 10.1128/spectrum.02405-21. Epub 2022 Oct 10. Microbiol Spectr. 2022. PMID: 36214695 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials