The yeast Red1 protein localizes to the cores of meiotic chromosomes - PubMed (original) (raw)
The yeast Red1 protein localizes to the cores of meiotic chromosomes
A V Smith et al. J Cell Biol. 1997.
Abstract
Mutants in the meiosis-specific RED1 gene of S. cerevisiae fail to make any synaptonemal complex (SC) or any obvious precursors to the SC. Using antibodies that specifically recognize the Red1 protein, Red1 has been localized along meiotic pachytene chromosomes. Red1 also localizes to the unsynapsed axial elements present in a zip1 mutant, suggesting that Red1 is a component of the lateral elements of mature SCs. Anti-Red1 staining is confined to the cores of meiotic chromosomes and is not associated with the loops of chromatin that lie outside the SC. Analysis of the spo11 mutant demonstrates that Red1 localization does not depend upon meiotic recombination. The localization of Red1 has been compared with two other meiosis-specific components of chromosomes, Hop1 and Zip1; Zip1 serves as a marker for synapsed chromosomes. Double labeling of wild-type meiotic chromosomes with anti-Zip1 and anti-Red1 antibodies demonstrates that Red1 localizes to chromosomes both before and during pachytene. Double labeling with anti-Hop1 and anti-Red1 antibodies reveals that Hop1 protein localizes only in areas that also contain Red1, and studies of Hop1 localization in a red1 null mutant demonstrate that Hop1 localization depends on Red1 function. These observations are consistent with previous genetic studies suggesting that Red1 and Hop1 directly interact. There is little or no Hop1 protein on pachytene chromosomes or in synapsed chromosomal regions.
Figures
Figure 1
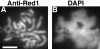
Red1 and Zip1 protein localization on wild-type chromosomes. Spread meiotic chromosomes from wild-type cells (BR2495) stained with mouse anti-Red1 antibodies (A, D, and G), rabbit anti-Zip1 antibodies (B, E, and H) and DAPI (C, F, and I). (A–C) Spread pachytene nucleus from 15-h time point. (D–F) Spread nucleus from 10-h time point. (G–I) Spread nucleus from 13-h time point. Categories 1, 2, and 3 are described in the text. Bar, 2 μm.
Figure 2
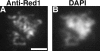
Red1 protein localization in strains overproducing Red1. Spread meiotic chromosomes from a wild-type cell overproducing Red1 (BS354) stained with rabbit anti-Red1 antibodies (A) and DAPI (B). Bar, 2 μm.
Figure 3
Red1 and histone localization following DNase I digestion of meiotic chromosomes. Wild-type spreads (BR2495) incubated for 1 h at 37°C with 0 U/ml (A–C) or 100 U/ml (D–F) DNase I. Spreads were stained with mouse anti-Red1 (A and D) and rabbit antihistone 2B (B and E) antibodies in addition to DAPI staining (C and F). Bar, 2 μm.
Figure 4
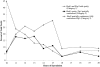
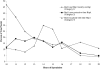
Red1 and Zip1 localization patterns during meiotic prophase progression. At each time point, 100 nuclei were examined and scored for Red1 and Zip1 localization patterns. Categories of staining are described in the text. The percentage of nuclei that stain with antibodies is always <100% because not all cells enter meiosis and those cells that undergo sporulation do not proceed through meiosis in synchrony.
Figure 5
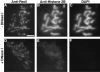
Red1 and Hop1 protein localization on wild-type chromosomes. Spread meiotic chromosomes from wild-type cells (BR2495) stained with mouse anti-Red1 antibodies (A, E, and I), rabbit anti-Hop1 antibodies (B, F, and J) and DAPI (D, H, and L). (A–D) Spread nucleus from 10-h time point with category 1 staining pattern. (E–H) Spread nucleus from 13-h time point with category 2 staining pattern. (I–L) Spread pachytene nucleus from 15-h time point with category 3 staining pattern. (C, G, and K) Fusion of anti-Red1 and anti-Hop1 staining patterns; areas of overlap appear as yellow spots. Bar, 2 μm.
Figure 6
Red1 and Hop1 localization patterns during meiotic prophase progression. At each time point, 100 nuclei were examined and scored for Red1 and Hop1 localization. Categories of staining are described in the text.
Figure 7
Hop1 and Zip1 protein localization on wild-type chromosomes. Spread zygotene nucleus from a wild-type cell (BR2495) from 13-h time point stained with rabbit anti-Hop1 antibodies (A) and mouse anti-Zip1 antibodies (B). Bar, 2 μm.
Figure 8
Red1 and Hop1 localization on zip1 chromosomes. Spread meiotic chromosomes from a zip1 cell (MY129) stained with mouse anti-Red1 antibodies (A), rabbit anti-Hop1 antibodies (B), and DAPI (C). Bar, 2 μm.
Figure 9
Zip1 localization on red1 chromosomes. Spread meiotic chromosomes from red1 cells (MY231) stained with rabbit antiZip1 antibodies (A) and DAPI (B). Bar, 2 μm.
Figure 10
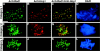
Red1 and Hop1 localization on spo11 chromosomes. Spread meiotic chromosomes from spo11 cells (S2888) stained with mouse anti-Red1 antibodies (A and D), rabbit anti-Hop1 antibodies (B and E), and DAPI (C and F). Bar, 2 μm.
Figure 11
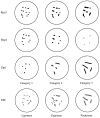
Summary of immunofluorescence and electron micrographic stages during meiotic prophase.
Similar articles
- A conditional allele of the Saccharomyces cerevisiae HOP1 gene is suppressed by overexpression of two other meiosis-specific genes: RED1 and REC104.
Hollingsworth NM, Johnson AD. Hollingsworth NM, et al. Genetics. 1993 Apr;133(4):785-97. doi: 10.1093/genetics/133.4.785. Genetics. 1993. PMID: 8462842 Free PMC article. - Bypass of a meiotic checkpoint by overproduction of meiotic chromosomal proteins.
Bailis JM, Smith AV, Roeder GS. Bailis JM, et al. Mol Cell Biol. 2000 Jul;20(13):4838-48. doi: 10.1128/MCB.20.13.4838-4848.2000. Mol Cell Biol. 2000. PMID: 10848609 Free PMC article. - Getting there: understanding the chromosomal recruitment of the AAA+ ATPase Pch2/TRIP13 during meiosis.
Cardoso da Silva R, Vader G. Cardoso da Silva R, et al. Curr Genet. 2021 Aug;67(4):553-565. doi: 10.1007/s00294-021-01166-3. Epub 2021 Mar 12. Curr Genet. 2021. PMID: 33712914 Free PMC article. Review. - Chromosome cores and chromatin at meiotic prophase.
Moens PB, Pearlman RE, Heng HH, Traut W. Moens PB, et al. Curr Top Dev Biol. 1998;37:241-62. doi: 10.1016/s0070-2153(08)60176-3. Curr Top Dev Biol. 1998. PMID: 9352188 Review.
Cited by
- Separable Crossover-Promoting and Crossover-Constraining Aspects of Zip1 Activity during Budding Yeast Meiosis.
Voelkel-Meiman K, Johnston C, Thappeta Y, Subramanian VV, Hochwagen A, MacQueen AJ. Voelkel-Meiman K, et al. PLoS Genet. 2015 Jun 26;11(6):e1005335. doi: 10.1371/journal.pgen.1005335. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26114667 Free PMC article. - Meiosis in mice without a synaptonemal complex.
Kouznetsova A, Benavente R, Pastink A, Höög C. Kouznetsova A, et al. PLoS One. 2011;6(12):e28255. doi: 10.1371/journal.pone.0028255. Epub 2011 Dec 2. PLoS One. 2011. PMID: 22164254 Free PMC article. - Prevention of DNA Rereplication Through a Meiotic Recombination Checkpoint Response.
Najor NA, Weatherford L, Brush GS. Najor NA, et al. G3 (Bethesda). 2016 Dec 7;6(12):3869-3881. doi: 10.1534/g3.116.033910. G3 (Bethesda). 2016. PMID: 27678521 Free PMC article. - Mek1 stabilizes Hop1-Thr318 phosphorylation to promote interhomolog recombination and checkpoint responses during yeast meiosis.
Chuang CN, Cheng YH, Wang TF. Chuang CN, et al. Nucleic Acids Res. 2012 Dec;40(22):11416-27. doi: 10.1093/nar/gks920. Epub 2012 Oct 9. Nucleic Acids Res. 2012. PMID: 23047948 Free PMC article. - Differential activation of M26-containing meiotic recombination hot spots in Schizosaccharomyces pombe.
Pryce DW, Lorenz A, Smirnova JB, Loidl J, McFarlane RJ. Pryce DW, et al. Genetics. 2005 May;170(1):95-106. doi: 10.1534/genetics.104.036301. Epub 2005 Mar 2. Genetics. 2005. PMID: 15744055 Free PMC article.
References
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. - PubMed
- Bishop DK. RecA homologues Dmc1 and Rad51 interact to form discrete nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. - PubMed
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. . Cell. 1990;61:1089–1101. - PubMed
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J Cell Sci. 1994;107:2749–2760. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases