Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited - PubMed (original) (raw)
Review
Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited
D Kolakofsky et al. J Virol. 1998 Feb.
No abstract available
Figures
FIG. 1
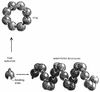
Single subunit assemblies. Large macromolecular assemblies can be formed from a single protein subunit, if the subunit interacts with itself repeatedly. This is possible if the binding site is complementary to a region of its own surface that does not include the binding site itself. Adapted from Alberts et al. (1).
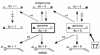
FIG. 2
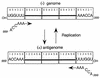
Paramyxovirus genome replication and the requirement for hexamer genome length. Genome and antigenome nucleocapsids are shown as a linear array of N subunits (open rectangles), each with six sites for binding nucleotides (numbered 1 to 6, 5′ to 3′ on the antigenome). Only the sequences of the six 5′ (ppp) and 3′ (OH) conserved bases are shown. Nucleocapsid assembly occurs concomitantly with synthesis and is proposed to initiate flush with the 5′ pppACCAAA hexanucleotide. Chains which are not 6n+0 long alter the position of the 3′ OHUGGUUU promoter element relative to the terminal N subunit and are therefore inefficiently initiated.
FIG. 3
_cis_-acting promoter sequences and the rule of six. The N-RNA nucleocapsid template is shown above as a helical assembly of N subunits, each containing 6-nt binding sites (dark ovals). The SeV sequence within the 9th, 10th, and 11th subunits, containing the N mRNA start site at nt 56, is shown below; the YARRGT repeats at nt 55 to 66 are highlighted in bold capitals. The effect of adding a total of 6 nt at nt 47 and 67 on the hexamer phase of these repeats is shown on the right, and their effect on the amplification activity of a DI genome in the absence of C protein expression (27) is shown on the left. The polymerase is postulated to recognize these ARRG repeats equally well when they are contained entirely within the 9th and 10th or 10th and 11th subunits, but these repeats are less well recognized when their phase relative to the N subunits is altered. Adapted from reference .
FIG. 4
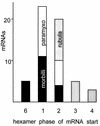
Distribution of mRNA start site subunit phases. The initiating adenosines of the 56 mRNA start sites listed in Table 1 are plotted in this histogram according to their N subunit positions (hexamer phase). The clustering of these phases for each genus of the subfamily is indicated by shading.
FIG. 5
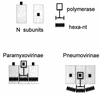
Paramyxoviridae and the rule of six. The N subunits are shown as open grey rectangles, with their six nucleotides shown as dark ovals. The polymerase binding site is shown as a small dark square, which is found at a different location relative to the RNA for each of the two subfamilies. The polymerase is shown as being composed of the N subunit binding site (open thick-lined rectangle) and the RNA interaction site (horizontal thick line) and is postulated to be similar for both subfamilies. The P and L components of the polymerase are not indicated. The simultaneous interaction of the polymerase N protein and RNA binding sites with their targets displaces the RNA from the N subunits for the pneumoviruses but does not disturb the RNA-N interactions for the paramyxoviruses.
FIG. 6
Paramyxovirus genome length correction. The viral RNAs are shown as horizontal lines; the filled symbols at the extremities indicate 3′ ends. The direction of RNA synthesis is indicated by arrows. Unnatural nonhexamer-length genomes (e.g., 6n+5) can be introduced into cells via T7 RNA polymerase expression vectors. Although they replicate poorly, their constant production by the bacteriophage polymerase ensures that some antigenome synthesis occurs. Purine insertions take place during antigenome synthesis as well as during mRNA synthesis, and those which are now of hexamer length replicate more efficiently and accumulate, leading to correction of the genome length.
Similar articles
- Paramyxovirus RNA editing and the requirement for hexamer genome length.
Hausmann S, Jacques JP, Kolakofsky D. Hausmann S, et al. RNA. 1996 Oct;2(10):1033-45. RNA. 1996. PMID: 8849779 Free PMC article. - Molecular basis for naturally occurring elevated readthrough transcription across the M-F junction of the paramyxovirus SV5.
Rassa JC, Parks GD. Rassa JC, et al. Virology. 1998 Aug 1;247(2):274-86. doi: 10.1006/viro.1998.9266. Virology. 1998. PMID: 9705920 - Viruses in the RNA world.
Rima BK. Rima BK. Biochem Soc Trans. 1996 Feb;24(1):1-13. doi: 10.1042/bst0240001. Biochem Soc Trans. 1996. PMID: 8674576 Review. No abstract available. - Paramyxovirus reverse genetics is coming of age.
Nagai Y, Kato A. Nagai Y, et al. Microbiol Immunol. 1999;43(7):613-24. doi: 10.1111/j.1348-0421.1999.tb02448.x. Microbiol Immunol. 1999. PMID: 10529101 Review. No abstract available.
Cited by
- Complete genome sequence and biological characterizations of a novel goose paramyxovirus-SF02 isolated in China.
Zou J, Shan S, Yao N, Gong Z. Zou J, et al. Virus Genes. 2005 Jan;30(1):13-21. doi: 10.1007/s11262-004-4577-x. Virus Genes. 2005. PMID: 15744558 - Full-length genome sequence and genetic relationship of two paramyxoviruses isolated from bat and pigs in the Americas.
Wang LF, Hansson E, Yu M, Chua KB, Mathe N, Crameri G, Rima BK, Moreno-López J, Eaton BT. Wang LF, et al. Arch Virol. 2007;152(7):1259-71. doi: 10.1007/s00705-007-0959-4. Epub 2007 Mar 26. Arch Virol. 2007. PMID: 17385069 Free PMC article. - RNA Synthesis and Capping by Non-segmented Negative Strand RNA Viral Polymerases: Lessons From a Prototypic Virus.
Ogino T, Green TJ. Ogino T, et al. Front Microbiol. 2019 Jul 10;10:1490. doi: 10.3389/fmicb.2019.01490. eCollection 2019. Front Microbiol. 2019. PMID: 31354644 Free PMC article. Review. - Complete genome sequences of avian paramyxovirus serotype 2 (APMV-2) strains Bangor, England and Kenya: evidence for the existence of subgroups within serotype 2.
Subbiah M, Nayak S, Collins PL, Samal SK. Subbiah M, et al. Virus Res. 2010 Sep;152(1-2):85-95. doi: 10.1016/j.virusres.2010.06.009. Epub 2010 Jun 22. Virus Res. 2010. PMID: 20600395 Free PMC article. - Whole genome sequencing and evolutionary analysis of human respiratory syncytial virus A and B from Milwaukee, WI 1998-2010.
Rebuffo-Scheer C, Bose M, He J, Khaja S, Ulatowski M, Beck ET, Fan J, Kumar S, Nelson MI, Henrickson KJ. Rebuffo-Scheer C, et al. PLoS One. 2011;6(10):e25468. doi: 10.1371/journal.pone.0025468. Epub 2011 Oct 6. PLoS One. 2011. PMID: 21998661 Free PMC article.
References
- Alberts B, et al. The molecular biology of the cell. 3rd ed. New York, N.Y: Garland Publishing; 1994.
- Baudin, F. (EMBL, Grenoble, France). Personal communication.
- Blumberg B M, Giorgi C, Kolakofsky D. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell. 1983;32:559–567. - PubMed
- Calain P, Curran J, Kolakofsky D, Roux L. Molecular cloning of natural paramyxovirus copy-back defective interfering RNAs and their expression from DNA. Virology. 1992;191:62–71. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources