Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes - PubMed (original) (raw)
Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes
A H Franks et al. Appl Environ Microbiol. 1998 Sep.
Abstract
Six 16S rRNA-targeted oligonucleotide probes were designed, validated, and used to quantify predominant groups of anaerobic bacteria in human fecal samples. A set of two probes was specific for species of the Bacteroides fragilis group and the species Bacteroides distasonis. Two others were designed to detect species of the Clostridium histolyticum and the Clostridium lituseburense groups. Another probe was designed for the genera Streptococcus and Lactococcus, and the final probe was designed for the species of the Clostridium coccoides-Eubacterium rectale group. The temperature of dissociation of each of the probes was determined. The specificities of the probes for a collection of target and reference organisms were tested by dot blot hybridization and fluorescent in situ hybridization (FISH). The new probes were used in initial FISH experiments to enumerate human fecal bacteria. The combination of the two Bacteroides-specific probes detected a mean of 5.4 x 10(10) cells per g (dry weight) of feces; the Clostridium coccoides-Eubacterium rectale group-specific probe detected a mean of 7.2 x 10(10) cells per g (dry weight) of feces. The Clostridium histolyticum, Clostridium lituseburense, and Streptococcus-Lactococcus group-specific probes detected only numbers of cells ranging from 1 x 10(7) to 7 x 10(8) per g (dry weight) of feces. Three of the newly designed probes and three additional probes were used in further FISH experiments to study the fecal flora composition of nine volunteers over a period of 8 months. The combination of probes was able to detect at least two-thirds of the fecal flora. The normal biological variations within the fecal populations of the volunteers were determined and indicated that these variations should be considered when evaluating the effects of agents modulating the flora.
Figures
FIG. 1
Alignments of the probe sequences, their target sites, and sequences of corresponding sites in reference organisms. Probe names are in accordance with the Oligonucleotide Probe Database nomenclature (1) and are abbreviated in the text. Only those nucleotides that are different from the target sequences are shown. Y is a (C/T) wobbled nucleotide and R is an (A/G) wobbled nucleotide. Bold underlined nucleotides match the wobbled nucleotide of the probe. C., Clostridium; B., Butyrivibrio. The numerals following some species names refer to sequence accession numbers.
FIG. 2
Phylogenetic trees illustrating the target groups of the new probes and related species. (A) Tree showing the relationship among the major genera, groups, and organisms known be present in the human gut. The heights of the boxes reflect the numbers of sequences used in the tree construction, and the horizontal lengths reflect the phylogenetic depths of the corresponding groups. Boxes and group names in boldface type indicate the groups that are represented in more detail in succeeding panels (B to F). The trees in panels B to F show the target organisms in bold italic typeface and related but nontarget organisms in normal typeface. (B) The complete genera Streptococcus and Lactococcus, all species of which are targets for the Strc493 probe. (C) The genus Bacteroides and related genera, with the target organisms for the Bfra602 and Bdis656 probes indicated. (D) Clostridium lituseburense group as a part of Clostridium cluster XI (classification according to reference 5), with the target organisms for the Clit135 probe indicated. (E) Clostridium histolyticum group (Clostridium clusters I and II), with the targets of the Chis150 probe indicated. (F) Clostridium coccoides-Eubacterium rectale group (Clostridium cluster CIV), with the targets for the Erec482 probe indicated. All bars represent 10% sequence divergence.
FIG. 2
Phylogenetic trees illustrating the target groups of the new probes and related species. (A) Tree showing the relationship among the major genera, groups, and organisms known be present in the human gut. The heights of the boxes reflect the numbers of sequences used in the tree construction, and the horizontal lengths reflect the phylogenetic depths of the corresponding groups. Boxes and group names in boldface type indicate the groups that are represented in more detail in succeeding panels (B to F). The trees in panels B to F show the target organisms in bold italic typeface and related but nontarget organisms in normal typeface. (B) The complete genera Streptococcus and Lactococcus, all species of which are targets for the Strc493 probe. (C) The genus Bacteroides and related genera, with the target organisms for the Bfra602 and Bdis656 probes indicated. (D) Clostridium lituseburense group as a part of Clostridium cluster XI (classification according to reference 5), with the target organisms for the Clit135 probe indicated. (E) Clostridium histolyticum group (Clostridium clusters I and II), with the targets of the Chis150 probe indicated. (F) Clostridium coccoides-Eubacterium rectale group (Clostridium cluster CIV), with the targets for the Erec482 probe indicated. All bars represent 10% sequence divergence.
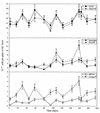
FIG. 3
Microbial composition of fecal samples of volunteer B. (Top panel) Total cells enumerated by DAPI staining and total bacteria counted by FISH with the Bact338 probe; (middle panel) Bacteroides cells enumerated with the Bfra602-Bdis656 probe combination and the Clostridium coccoides-Eubacterium rectale group cells enumerated with the Erec482 probe; (bottom panel) bifidobacteria and the Low G+C #2 group enumerated with the Bif164 probe and the Lowgc2P probe, respectively.
Similar articles
- Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization.
Sghir A, Gramet G, Suau A, Rochet V, Pochart P, Dore J. Sghir A, et al. Appl Environ Microbiol. 2000 May;66(5):2263-6. doi: 10.1128/AEM.66.5.2263-2266.2000. Appl Environ Microbiol. 2000. PMID: 10788414 Free PMC article. - Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacterium rectale.
Aminov RI, Walker AW, Duncan SH, Harmsen HJ, Welling GW, Flint HJ. Aminov RI, et al. Appl Environ Microbiol. 2006 Sep;72(9):6371-6. doi: 10.1128/AEM.00701-06. Appl Environ Microbiol. 2006. PMID: 16957265 Free PMC article. - A survey of the relative abundance of specific groups of cellulose degrading bacteria in anaerobic environments using fluorescence in situ hybridization.
O'Sullivan C, Burrell PC, Clarke WP, Blackall LL. O'Sullivan C, et al. J Appl Microbiol. 2007 Oct;103(4):1332-43. doi: 10.1111/j.1365-2672.2007.03362.x. J Appl Microbiol. 2007. PMID: 17897237 Review.
Cited by
- Use of 16S rRNA Gene-Targeted Group-Specific Primers for Real-Time PCR Analysis of Predominant Bacteria in Mouse Feces.
Yang YW, Chen MK, Yang BY, Huang XJ, Zhang XR, He LQ, Zhang J, Hua ZC. Yang YW, et al. Appl Environ Microbiol. 2015 Oct;81(19):6749-56. doi: 10.1128/AEM.01906-15. Epub 2015 Jul 17. Appl Environ Microbiol. 2015. PMID: 26187967 Free PMC article. - The impact of date palm fruits and their component polyphenols, on gut microbial ecology, bacterial metabolites and colon cancer cell proliferation.
Eid N, Enani S, Walton G, Corona G, Costabile A, Gibson G, Rowland I, Spencer JP. Eid N, et al. J Nutr Sci. 2014 Oct 8;3:e46. doi: 10.1017/jns.2014.16. eCollection 2014. J Nutr Sci. 2014. PMID: 26101614 Free PMC article. - Crohn's disease patients have more IgG-binding fecal bacteria than controls.
Harmsen HJ, Pouwels SD, Funke A, Bos NA, Dijkstra G. Harmsen HJ, et al. Clin Vaccine Immunol. 2012 Apr;19(4):515-21. doi: 10.1128/CVI.05517-11. Epub 2012 Feb 15. Clin Vaccine Immunol. 2012. PMID: 22336288 Free PMC article. - Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples.
Ahmed S, Macfarlane GT, Fite A, McBain AJ, Gilbert P, Macfarlane S. Ahmed S, et al. Appl Environ Microbiol. 2007 Nov;73(22):7435-42. doi: 10.1128/AEM.01143-07. Epub 2007 Sep 21. Appl Environ Microbiol. 2007. PMID: 17890331 Free PMC article. - Clinical, microbiological, and immunological effects of fructo-oligosaccharide in patients with Crohn's disease.
Lindsay JO, Whelan K, Stagg AJ, Gobin P, Al-Hassi HO, Rayment N, Kamm MA, Knight SC, Forbes A. Lindsay JO, et al. Gut. 2006 Mar;55(3):348-55. doi: 10.1136/gut.2005.074971. Epub 2005 Sep 14. Gut. 2006. PMID: 16162680 Free PMC article. Clinical Trial.
References
- Borriello, S. P. 1995. Clostridial disease of the gut. Clin. Infect. Dis. 20(Suppl. 2):S242–S250. - PubMed
- Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical