ERBB2, CPTAC-192 - CPTAC Assay Portal (original) (raw)
Please include the following statement when referencing the CPTAC Assay Portal
We would like to acknowledge the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) Assay Portal (assays.cancer.gov) for developing assays and establishing criteria for the assays described in this publication.
Overview Data source: UniProt
| Official Gene Symbol | Other Aliases |
|---|---|
| ERBB2 | HER2, MLN19, NEU, NGL |
| Sequence Length (AA) | Molecular Weight (Da) |
|---|---|
| 1255 | 137910 |
| Protein Name |
|---|
| Receptor tyrosine-protein kinase erbB-2 |
| Sources | |
|---|---|
| UniProt PhosphoSitePlus ® GeneCards | Human Protein Atlas |
Protein Sequence hover to view complete sequence
CPTAC-194: View additional ELVSEFSR data
non-CPTAC-5385: View additional ELVSEFSR data
non-CPTAC-5385: View additional ELVSEFSR data
CPTAC-5777: View additional GIWIPDGENVK data
non-CPTAC-5547: View additional GIWIPDGENVK data
| 10 | 20 | 30 | 40 | 50 |
|---|---|---|---|---|
| MELAALCRWG | LLLALLPPGA | ASTQVCTGTD | MKLRLPASPE | THLDMLRHLY |
| 60 | 70 | 80 | 90 | 100 |
| QGCQVVQGNL | ELTYLPTNAS | LSFLQDIQEV | QGYVLIAHNQ | VRQVPLQRLR |
| 110 | 120 | 130 | 140 | 150 |
| IVRGTQLFED | NYALAVLDNG | DPLNNTTPVT | GASPGGLREL | QLRSLTEILK |
| 160 | 170 | 180 | 190 | 200 |
| GGVLIQRNPQ | LCYQDTILWK | DIFHKNNQLA | LTLIDTNRSR | ACHPCSPMCK |
| 210 | 220 | 230 | 240 | 250 |
| GSRCWGESSE | DCQSLTRTVC | AGGCARCKGP | LPTDCCHEQC | AAGCTGPKHS |
| 260 | 270 | 280 | 290 | 300 |
| DCLACLHFNH | SGICELHCPA | LVTYNTDTFE | SMPNPEGRYT | FGASCVTACP |
| 310 | 320 | 330 | 340 | 350 |
| YNYLSTDVGS | CTLVCPLHNQ | EVTAEDGTQR | CEKCSKPCAR | VCYGLGMEHL |
| 360 | 370 | 380 | 390 | 400 |
| REVRAVTSAN | IQEFAGCKKI | FGSLAFLPES | FDGDPASNTA | PLQPEQLQVF |
| 410 | 420 | 430 | 440 | 450 |
| ETLEEITGYL | YISAWPDSLP | DLSVFQNLQV | IRGRILHNGA | YSLTLQGLGI |
| 460 | 470 | 480 | 490 | 500 |
| SWLGLRSLRE | LGSGLALIHH | NTHLCFVHTV | PWDQLFRNPH | QALLHTANRP |
| 510 | 520 | 530 | 540 | 550 |
| EDECVGEGLA | CHQLCARGHC | WGPGPTQCVN | CSQFLRGQEC | VEECRVLQGL |
| 560 | 570 | 580 | 590 | 600 |
| PREYVNARHC | LPCHPECQPQ | NGSVTCFGPE | ADQCVACAHY | KDPPFCVARC |
| 610 | 620 | 630 | 640 | 650 |
| PSGVKPDLSY | MPIWKFPDEE | GACQPCPINC | THSCVDLDDK | GCPAEQRASP |
| 660 | 670 | 680 | 690 | 700 |
| LTSIISAVVG | ILLVVVLGVV | FGILIKRRQQ | KIRKYTMRRL | LQETELVEPL |
| 710 | 720 | 730 | 740 | 750 |
| TPSGAMPNQA | QMRILKETEL | RKVKVLGSGA | FGTVYKGIWI | PDGENVKIPV |
| 760 | 770 | 780 | 790 | 800 |
| AIKVLRENTS | PKANKEILDE | AYVMAGVGSP | YVSRLLGICL | TSTVQLVTQL |
| 810 | 820 | 830 | 840 | 850 |
| MPYGCLLDHV | RENRGRLGSQ | DLLNWCMQIA | KGMSYLEDVR | LVHRDLAARN |
| 860 | 870 | 880 | 890 | 900 |
| VLVKSPNHVK | ITDFGLARLL | DIDETEYHAD | GGKVPIKWMA | LESILRRRFT |
| 910 | 920 | 930 | 940 | 950 |
| HQSDVWSYGV | TVWELMTFGA | KPYDGIPARE | IPDLLEKGER | LPQPPICTID |
| 960 | 970 | 980 | 990 | 1000 |
| VYMIMVKCWM | IDSECRPRFR | ELVSEFSRMA | RDPQRFVVIQ | NEDLGPASPL |
| 1010 | 1020 | 1030 | 1040 | 1050 |
| DSTFYRSLLE | DDDMGDLVDA | EEYLVPQQGF | FCPDPAPGAG | GMVHHRHRSS |
| 1060 | 1070 | 1080 | 1090 | 1100 |
| STRSGGGDLT | LGLEPSEEEA | PRSPLAPSEG | AGSDVFDGDL | GMGAAKGLQS |
| 1110 | 1120 | 1130 | 1140 | 1150 |
| LPTHDPSPLQ | RYSEDPTVPL | PSETDGYVAP | LTCSPQPEYV | NQPDVRPQPP |
| 1160 | 1170 | 1180 | 1190 | 1200 |
| SPREGPLPAA | RPAGATLERP | KTLSPGKNGV | VKDVFAFGGA | VENPEYLTPQ |
| 1210 | 1220 | 1230 | 1240 | 1250 |
| GGAAPQPHPP | PAFSPAFDNL | YYWDQDPPER | GAPPSTFKGT | PTAENPEYLG |
| 1255 | ||||
| LDVPV |
Data source: UniProt
Position of Targeted Peptide Analytes Relative to SNPs, Isoforms, and PTMs
Uniprot Database Entry PhosphoSitePlus ®
Click a point on a node
to view detailed assay information below
All other points link out to UniProt
Phosphorylation Acetylation Ubiquitylation Other
loading

Assay Details for CPTAC-194 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
ELVSEFSR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
971
Peptide End
978
CPTAC ID
CPTAC-194
Peptide Molecular Mass
965.4818
Species
Homo sapiens (Human)
Assay Type
Direct MRM
Matrix
cell line lysate pool
Submitting Laboratory
Fred Hutchinson Cancer Research Center
Submitting Lab PI
Amanda Paulovich
Publication
View Details (opens in a new window)
Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Kennedy JJ, Abbatiello SE, Kim K, Yan P, Whiteaker JR, Lin C, Kim JS, Zhang Y, Wang X, Ivey RG, Zhao L, Min H, Lee Y, Yu MH, Yang EG, Lee C, Wang P, Rodriguez H, Kim Y, Carr SA, Paulovich AG. Nat Methods. 2014 Feb;11(2):149-55. doi: 10.1038/nmeth.2763. Epub 2013 Dec 8. PMID: 24317253
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
5500 QTRAP (ABSCIEX)
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C,15N
LC
nanoLC-Ultra 2D, cHiPLC-nanoflex (Eksigent)
Column Packing
ChromXP C18-CL, 3 um, 120A
Column Dimensions
150 x 0.075 mm
Flow Rate
500 nL / min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

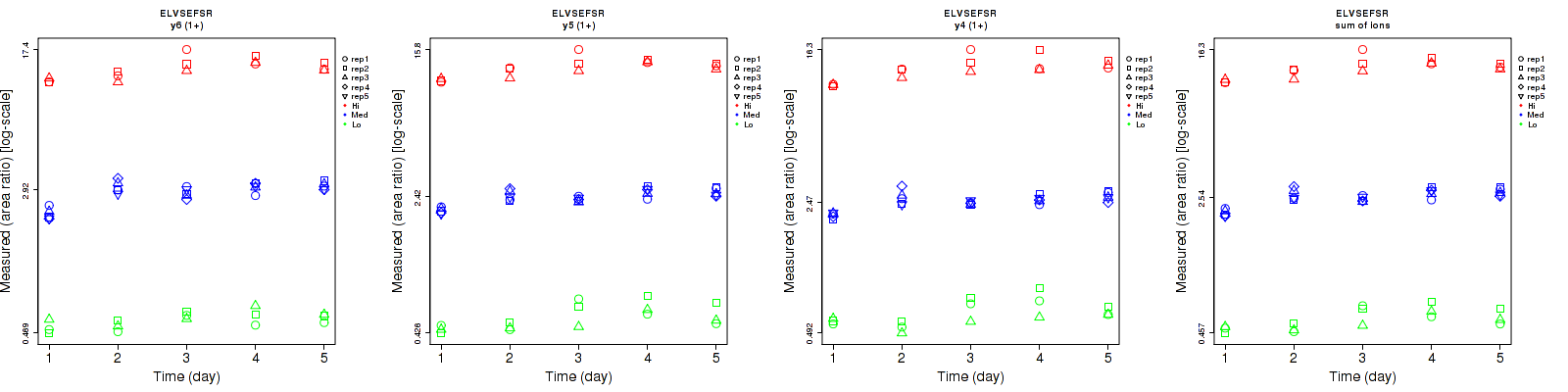
Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y6 (1+) | 7.8 | 6.6 | 6.8 | 9.4 | 15 | 12.8 | 12.2 | 16.4 | 14.5 | 15 | 25 | 15 |
| y5 (1+) | 10.9 | 5 | 5.7 | 15.3 | 10.8 | 11.2 | 18.8 | 11.9 | 12.6 | 15 | 25 | 15 |
| y4 (1+) | 9.7 | 5.1 | 8 | 13.6 | 10.5 | 13.5 | 16.7 | 11.7 | 15.7 | 15 | 25 | 15 |
| sum | 8.5 | 5.2 | 6.2 | 12.9 | 11.7 | 11.8 | 15.4 | 12.8 | 13.3 | 15 | 25 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y6 (1+) | 7.8 | 6.6 | 6.8 | 9.4 | 15 | 12.8 | 12.2 | 16.4 | 14.5 | 15 | 25 | 15 |
| y5 (1+) | 10.9 | 5 | 5.7 | 15.3 | 10.8 | 11.2 | 18.8 | 11.9 | 12.6 | 15 | 25 | 15 |
| y4 (1+) | 9.7 | 5.1 | 8 | 13.6 | 10.5 | 13.5 | 16.7 | 11.7 | 15.7 | 15 | 25 | 15 |
| sum | 8.5 | 5.2 | 6.2 | 12.9 | 11.7 | 11.8 | 15.4 | 12.8 | 13.3 | 15 | 25 | 15 |
Additional Resources and Comments
Assay Details for CPTAC-5864 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
GLQSLPTHDPSPLQR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
6
Peptide Start
1097
Peptide End
1111
CPTAC ID
CPTAC-5864
Peptide Molecular Mass
1,644.8584
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
N/A
Matrix
FFPE breast cancer tumor tissue lysate pool
Submitting Laboratory
Fred Hutchinson Cancer Research Center
Submitting Lab PI
Amanda Paulovich
Publication
View Details (opens in a new window)
Quantification of Human Epidermal Growth Factor Receptor 2 by Immunopeptide Enrichment and Targeted Mass Spectrometry in Formalin-Fixed Paraffin-Embedded and Frozen Breast Cancer Tissues. Kennedy JJ, Whiteaker JR, Kennedy LC, Bosch DE, Lerch ML, Schoenherr RM, Zhao L, Lin C, Chowdhury S, Kilgore MR, Allison KH, Wang P, Hoofnagle AN, Baird GS, Paulovich AG. Clin Chem. 2021 Jun 17:hvab047. doi: 10.1093/clinchem/hvab047. Online ahead of print. PMID: 34136904
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
QTRAP 6500
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus R
LC
Eksigent nanoLC 425
Column Packing
3 µm ChromXP C18EP, 120 Å
Column Dimensions
150 x 0.3 mm
Flow Rate
10.0 µL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

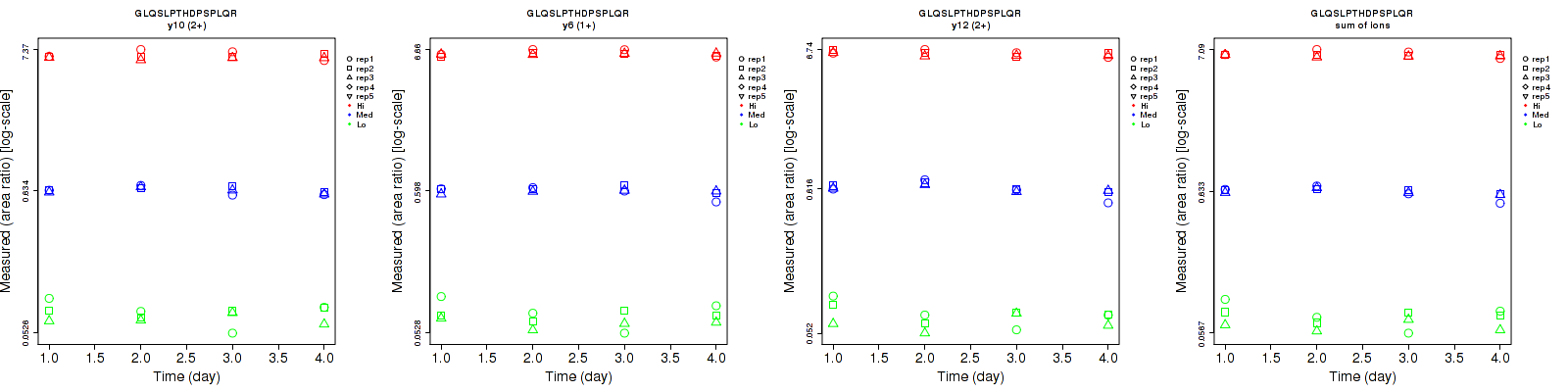
Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y12 (2+) | 16.2 | 5.2 | 3.9 | 17.2 | 9.3 | 4.3 | 23.6 | 10.7 | 5.8 | 12 | 12 | 12 |
| y13 (2+) | 22.2 | 9.7 | 3.8 | 16.8 | 12.8 | 3.6 | 27.8 | 16.1 | 5.2 | 12 | 12 | 12 |
| y6 (1+) | 17.3 | 5.9 | 3.6 | 13.4 | 6.2 | 3.6 | 21.9 | 8.6 | 5.1 | 12 | 12 | 12 |
| y10 (2+) | 16 | 3.8 | 5.5 | 13.2 | 5.9 | 4.3 | 20.7 | 7 | 7 | 12 | 12 | 12 |
| sum | 16.7 | 4.2 | 3.6 | 13.5 | 7 | 3 | 21.5 | 8.2 | 4.7 | 12 | 12 | 12 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y12 (2+) | 16.2 | 5.2 | 3.9 | 17.2 | 9.3 | 4.3 | 23.6 | 10.7 | 5.8 | 12 | 12 | 12 |
| y13 (2+) | 22.2 | 9.7 | 3.8 | 16.8 | 12.8 | 3.6 | 27.8 | 16.1 | 5.2 | 12 | 12 | 12 |
| y6 (1+) | 17.3 | 5.9 | 3.6 | 13.4 | 6.2 | 3.6 | 21.9 | 8.6 | 5.1 | 12 | 12 | 12 |
| y10 (2+) | 16 | 3.8 | 5.5 | 13.2 | 5.9 | 4.3 | 20.7 | 7 | 7 | 12 | 12 | 12 |
| sum | 16.7 | 4.2 | 3.6 | 13.5 | 7 | 3 | 21.5 | 8.2 | 4.7 | 12 | 12 | 12 |
Additional Resources and Comments
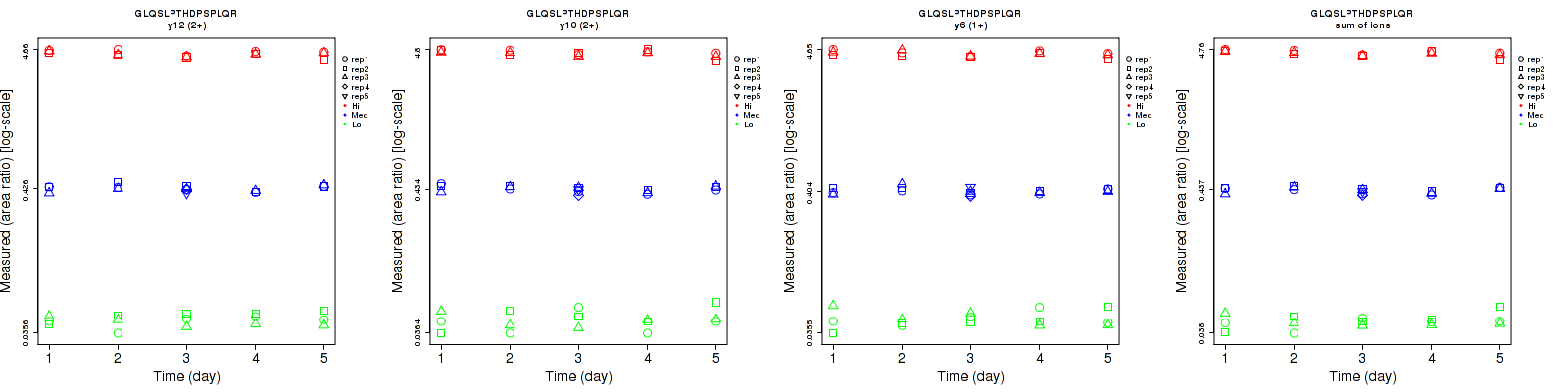
Assay Details for CPTAC-195 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
GLQSLPTHDPSPLQR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
1097
Peptide End
1111
CPTAC ID
CPTAC-195
Peptide Molecular Mass
1,644.8584
Species
Homo sapiens (Human)
Assay Type
Direct MRM
Matrix
cell line lysate pool
Submitting Laboratory
Fred Hutchinson Cancer Research Center
Submitting Lab PI
Amanda Paulovich
Publication
View Details (opens in a new window)
Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Kennedy JJ, Abbatiello SE, Kim K, Yan P, Whiteaker JR, Lin C, Kim JS, Zhang Y, Wang X, Ivey RG, Zhao L, Min H, Lee Y, Yu MH, Yang EG, Lee C, Wang P, Rodriguez H, Kim Y, Carr SA, Paulovich AG. Nat Methods. 2014 Feb;11(2):149-55. doi: 10.1038/nmeth.2763. Epub 2013 Dec 8. PMID: 24317253
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
5500 QTRAP (ABSCIEX)
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus R
LC
nanoLC-Ultra 2D, cHiPLC-nanoflex (Eksigent)
Column Packing
ChromXP C18-CL, 3 um, 120A
Column Dimensions
150 x 0.075 mm
Flow Rate
500 nL / min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

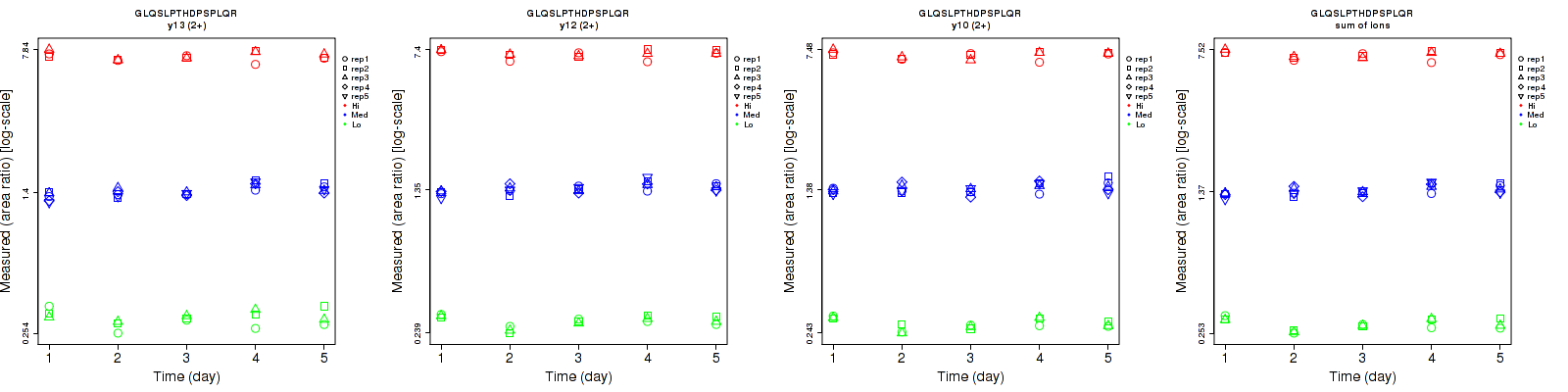
Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y13 (2+) | 8 | 4.2 | 3.6 | 8.8 | 6.9 | 4.8 | 11.9 | 8.1 | 6 | 15 | 25 | 15 |
| y12 (2+) | 3.3 | 4.4 | 3.7 | 6.9 | 5.9 | 4.3 | 7.6 | 7.4 | 5.7 | 15 | 25 | 15 |
| y10 (2+) | 3.7 | 5.3 | 3.2 | 6.7 | 6.1 | 4.4 | 7.7 | 8.1 | 5.4 | 15 | 25 | 15 |
| sum | 3.5 | 4.2 | 3.1 | 6.8 | 5.8 | 4.2 | 7.6 | 7.2 | 5.2 | 15 | 25 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y13 (2+) | 8 | 4.2 | 3.6 | 8.8 | 6.9 | 4.8 | 11.9 | 8.1 | 6 | 15 | 25 | 15 |
| y12 (2+) | 3.3 | 4.4 | 3.7 | 6.9 | 5.9 | 4.3 | 7.6 | 7.4 | 5.7 | 15 | 25 | 15 |
| y10 (2+) | 3.7 | 5.3 | 3.2 | 6.7 | 6.1 | 4.4 | 7.7 | 8.1 | 5.4 | 15 | 25 | 15 |
| sum | 3.5 | 4.2 | 3.1 | 6.8 | 5.8 | 4.2 | 7.6 | 7.2 | 5.2 | 15 | 25 | 15 |
Additional Resources and Comments
Assay Details for CPTAC-693 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
GLQSLPTHDPSPLQR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
1097
Peptide End
1111
CPTAC ID
CPTAC-693
Peptide Molecular Mass
1,644.8584
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
N/A
Matrix
plasma
Submitting Laboratory
Fred Hutchinson Cancer Research Center
Submitting Lab PI
Amanda Paulovich
Publication
View Details (opens in a new window)
Anti-peptide monoclonal antibodies generated for immuno-multiple reaction monitoring-mass spectrometry assays have a high probability of supporting Western blot and ELISA. Schoenherr RM, Saul RG, Whiteaker JR, Yan P, Whiteley GR, Paulovich AG. Mol Cell Proteomics. 2015 Feb;14(2):382-98. doi: 10.1074/mcp.O114.043133. Epub 2014 Dec 15. PMID: 25512614
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
5500 Qtrap
Internal Standard
synthetic peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus R
LC
Eksigent NanoLC 2D
Column Packing
ReproSil-Pur C18-AQ 3 um
Column Dimensions
100 mm x 75 um ID
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

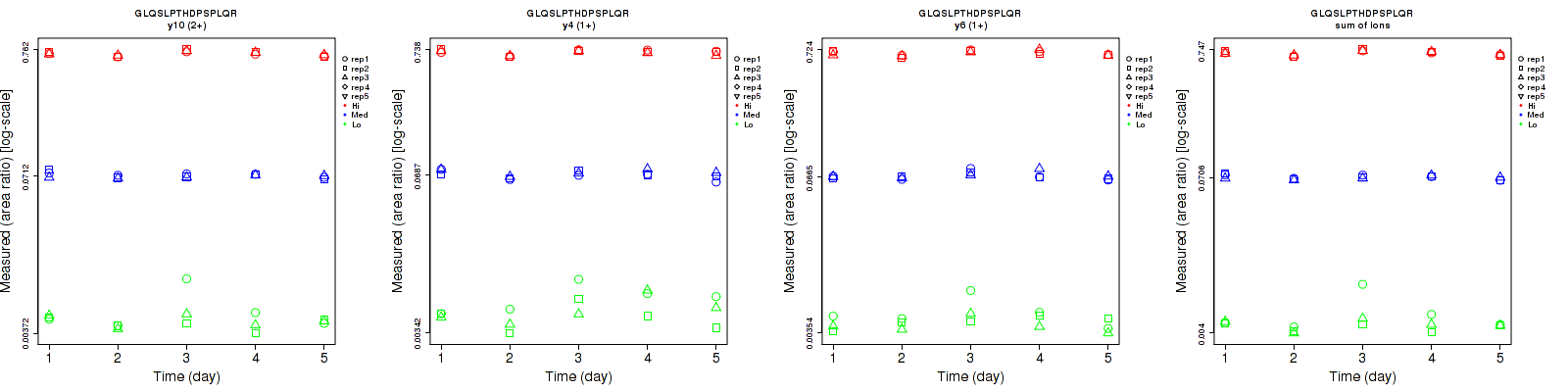
Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y10 (2+) | 15.4 | 3.5 | 1.9 | 21.9 | 4.1 | 4.4 | 26.8 | 5.4 | 4.8 | 15 | 15 | 15 |
| y4 (1+) | 22.6 | 5.5 | 2.4 | 26.3 | 6.8 | 4.9 | 34.7 | 8.7 | 5.5 | 15 | 15 | 15 |
| y6 (1+) | 16.8 | 4.9 | 2.8 | 18 | 6.9 | 5 | 24.6 | 8.5 | 5.7 | 15 | 15 | 15 |
| sum | 13.6 | 2.8 | 1.4 | 18.5 | 4.2 | 4.3 | 23 | 5 | 4.5 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y10 (2+) | 15.4 | 3.5 | 1.9 | 21.9 | 4.1 | 4.4 | 26.8 | 5.4 | 4.8 | 15 | 15 | 15 |
| y4 (1+) | 22.6 | 5.5 | 2.4 | 26.3 | 6.8 | 4.9 | 34.7 | 8.7 | 5.5 | 15 | 15 | 15 |
| y6 (1+) | 16.8 | 4.9 | 2.8 | 18 | 6.9 | 5 | 24.6 | 8.5 | 5.7 | 15 | 15 | 15 |
| sum | 13.6 | 2.8 | 1.4 | 18.5 | 4.2 | 4.3 | 23 | 5 | 4.5 | 15 | 15 | 15 |
Additional Resources and Comments
Assay Details for CPTAC-5839 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
GLQSLPTHDPSPLQR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
6
Peptide Start
1097
Peptide End
1111
CPTAC ID
CPTAC-5839
Peptide Molecular Mass
1,644.8584
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
N/A
Matrix
Frozen breast cancer tumor tissue lysate pool
Submitting Laboratory
Fred Hutchinson Cancer Research Center
Submitting Lab PI
Amanda Paulovich
Publication
View Details (opens in a new window)
Quantification of Human Epidermal Growth Factor Receptor 2 by Immunopeptide Enrichment and Targeted Mass Spectrometry in Formalin-Fixed Paraffin-Embedded and Frozen Breast Cancer Tissues. Kennedy JJ, Whiteaker JR, Kennedy LC, Bosch DE, Lerch ML, Schoenherr RM, Zhao L, Lin C, Chowdhury S, Kilgore MR, Allison KH, Wang P, Hoofnagle AN, Baird GS, Paulovich AG. Clin Chem. 2021 Jun 17:hvab047. doi: 10.1093/clinchem/hvab047. Online ahead of print. PMID: 34136904
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
QTRAP 6500
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus R
LC
Eksigent nanoLC 425
Column Packing
3 µm ChromXP C18EP, 120 Å
Column Dimensions
150 x 0.3 mm
Flow Rate
10.0 µL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y6 (1+) | 14.6 | 4.6 | 3.3 | 15 | 4.9 | 4.1 | 20.9 | 6.7 | 5.3 | 15 | 17 | 15 |
| y13 (2+) | 12.8 | 6.8 | 3.5 | 16.9 | 8.1 | 5.3 | 21.2 | 10.6 | 6.4 | 15 | 17 | 15 |
| y12 (2+) | 11.1 | 3.8 | 3.7 | 8.9 | 5.1 | 4.1 | 14.2 | 6.4 | 5.5 | 15 | 17 | 15 |
| y10 (2+) | 17.4 | 4.4 | 3.7 | 16.6 | 4.9 | 4.8 | 24 | 6.6 | 6.1 | 15 | 17 | 15 |
| sum | 11.5 | 3.3 | 2.7 | 11.5 | 4.6 | 4.1 | 16.3 | 5.7 | 4.9 | 15 | 17 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y6 (1+) | 14.6 | 4.6 | 3.3 | 15 | 4.9 | 4.1 | 20.9 | 6.7 | 5.3 | 15 | 17 | 15 |
| y13 (2+) | 12.8 | 6.8 | 3.5 | 16.9 | 8.1 | 5.3 | 21.2 | 10.6 | 6.4 | 15 | 17 | 15 |
| y12 (2+) | 11.1 | 3.8 | 3.7 | 8.9 | 5.1 | 4.1 | 14.2 | 6.4 | 5.5 | 15 | 17 | 15 |
| y10 (2+) | 17.4 | 4.4 | 3.7 | 16.6 | 4.9 | 4.8 | 24 | 6.6 | 6.1 | 15 | 17 | 15 |
| sum | 11.5 | 3.3 | 2.7 | 11.5 | 4.6 | 4.1 | 16.3 | 5.7 | 4.9 | 15 | 17 | 15 |
Additional Resources and Comments
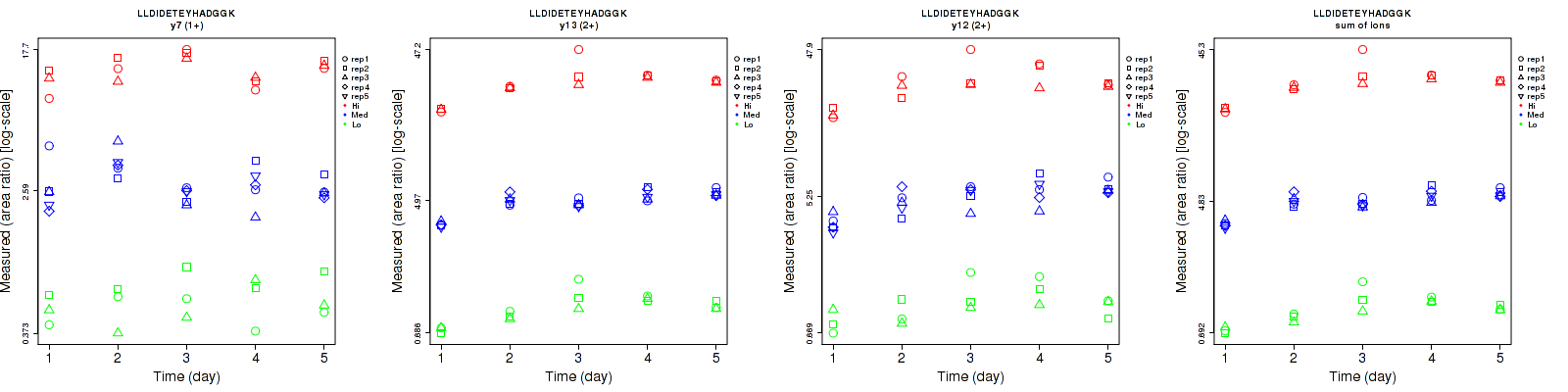
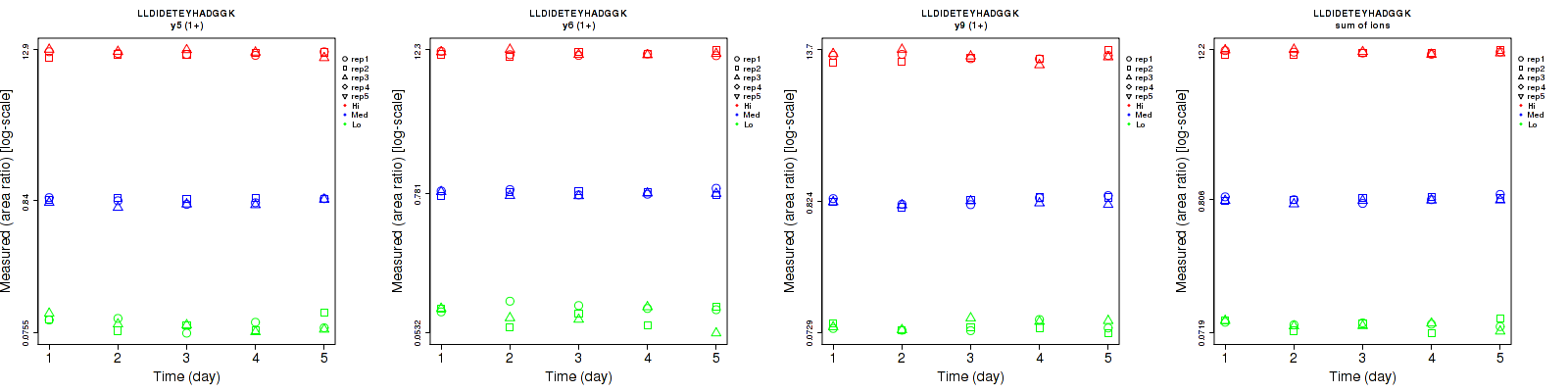
Assay Details for CPTAC-193 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
LLDIDETEYHADGGK
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
869
Peptide End
883
CPTAC ID
CPTAC-193
Peptide Molecular Mass
1,674.7737
Species
Homo sapiens (Human)
Assay Type
Direct MRM
Matrix
cell line lysate pool
Submitting Laboratory
Fred Hutchinson Cancer Research Center
Submitting Lab PI
Amanda Paulovich
Publication
View Details (opens in a new window)
Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Kennedy JJ, Abbatiello SE, Kim K, Yan P, Whiteaker JR, Lin C, Kim JS, Zhang Y, Wang X, Ivey RG, Zhao L, Min H, Lee Y, Yu MH, Yang EG, Lee C, Wang P, Rodriguez H, Kim Y, Carr SA, Paulovich AG. Nat Methods. 2014 Feb;11(2):149-55. doi: 10.1038/nmeth.2763. Epub 2013 Dec 8. PMID: 24317253
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
5500 QTRAP (ABSCIEX)
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus K
LC
nanoLC-Ultra 2D, cHiPLC-nanoflex (Eksigent)
Column Packing
ChromXP C18-CL, 3 um, 120A
Column Dimensions
150 x 0.075 mm
Flow Rate
500 nL / min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 29.9 | 22.1 | 10.9 | 21.6 | 28.9 | 18.4 | 36.9 | 36.4 | 21.4 | 15 | 25 | 15 |
| y13 (2+) | 8.5 | 6.1 | 7.3 | 21.4 | 18.2 | 22.9 | 23 | 19.2 | 24 | 15 | 25 | 15 |
| y12 (2+) | 20.7 | 15.2 | 15.4 | 23.7 | 23.5 | 25.6 | 31.5 | 28 | 29.9 | 15 | 25 | 15 |
| sum | 8.3 | 6.8 | 7.9 | 20.4 | 18 | 22.3 | 22 | 19.2 | 23.7 | 15 | 25 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 29.9 | 22.1 | 10.9 | 21.6 | 28.9 | 18.4 | 36.9 | 36.4 | 21.4 | 15 | 25 | 15 |
| y13 (2+) | 8.5 | 6.1 | 7.3 | 21.4 | 18.2 | 22.9 | 23 | 19.2 | 24 | 15 | 25 | 15 |
| y12 (2+) | 20.7 | 15.2 | 15.4 | 23.7 | 23.5 | 25.6 | 31.5 | 28 | 29.9 | 15 | 25 | 15 |
| sum | 8.3 | 6.8 | 7.9 | 20.4 | 18 | 22.3 | 22 | 19.2 | 23.7 | 15 | 25 | 15 |
Additional Resources and Comments
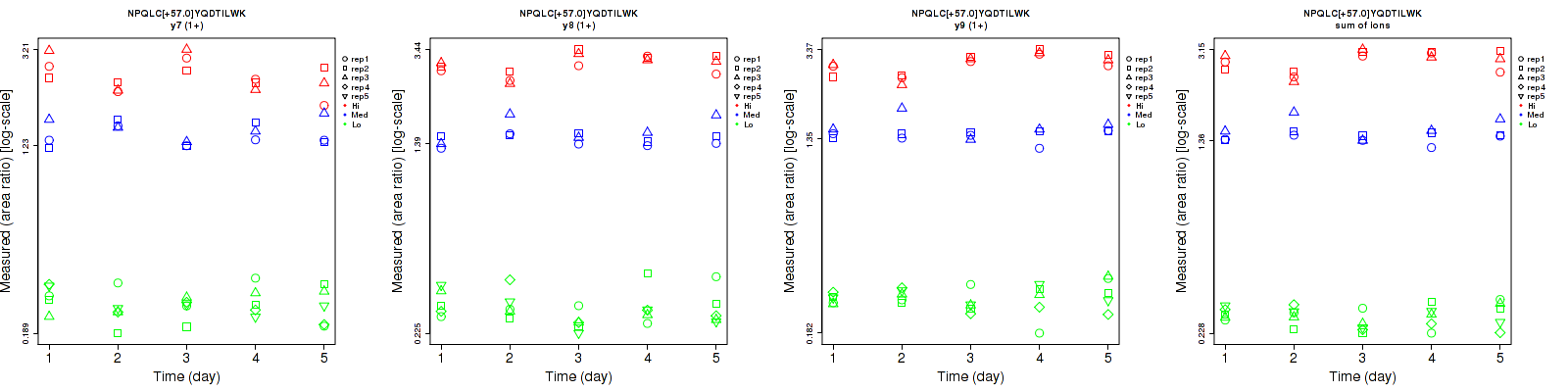
Assay Details for CPTAC-192 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Modified Sequence
NPQLC[+57.0]YQDTILWK
Modification Type
Carbamidomethyl Cysteine
Protein - Site of Modification
162
Peptide - Site of Modification
5
Peptide Start
158
Peptide End
170
CPTAC ID
CPTAC-192
Peptide Molecular Mass
1,677.8185
Species
Homo sapiens (Human)
Assay Type
Direct MRM
Matrix
cell line lysate pool
Submitting Laboratory
Fred Hutchinson Cancer Research Center
Submitting Lab PI
Amanda Paulovich
Publication
View Details (opens in a new window)
Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Kennedy JJ, Abbatiello SE, Kim K, Yan P, Whiteaker JR, Lin C, Kim JS, Zhang Y, Wang X, Ivey RG, Zhao L, Min H, Lee Y, Yu MH, Yang EG, Lee C, Wang P, Rodriguez H, Kim Y, Carr SA, Paulovich AG. Nat Methods. 2014 Feb;11(2):149-55. doi: 10.1038/nmeth.2763. Epub 2013 Dec 8. PMID: 24317253
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
5500 QTRAP (ABSCIEX)
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C,15N
LC
nanoLC-Ultra 2D, cHiPLC-nanoflex (Eksigent)
Column Packing
ChromXP C18-CL, 3 um, 120A
Column Dimensions
150 x 0.075 mm
Flow Rate
500 nL / min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 15.2 | 9.4 | 10.7 | 15.3 | 10.7 | 15.6 | 21.6 | 14.2 | 18.9 | 25 | 15 | 15 |
| y8 (1+) | 15.5 | 8.7 | 5.7 | 17.3 | 7.1 | 9.2 | 23.2 | 11.2 | 10.8 | 25 | 15 | 15 |
| y9 (1+) | 11.6 | 8 | 4.4 | 12.4 | 7.2 | 11 | 17 | 10.8 | 11.8 | 25 | 15 | 15 |
| sum | 9.3 | 7.3 | 5.4 | 10.1 | 5.9 | 9.9 | 13.7 | 9.4 | 11.3 | 25 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 15.2 | 9.4 | 10.7 | 15.3 | 10.7 | 15.6 | 21.6 | 14.2 | 18.9 | 25 | 15 | 15 |
| y8 (1+) | 15.5 | 8.7 | 5.7 | 17.3 | 7.1 | 9.2 | 23.2 | 11.2 | 10.8 | 25 | 15 | 15 |
| y9 (1+) | 11.6 | 8 | 4.4 | 12.4 | 7.2 | 11 | 17 | 10.8 | 11.8 | 25 | 15 | 15 |
| sum | 9.3 | 7.3 | 5.4 | 10.1 | 5.9 | 9.9 | 13.7 | 9.4 | 11.3 | 25 | 15 | 15 |
Additional Resources and Comments
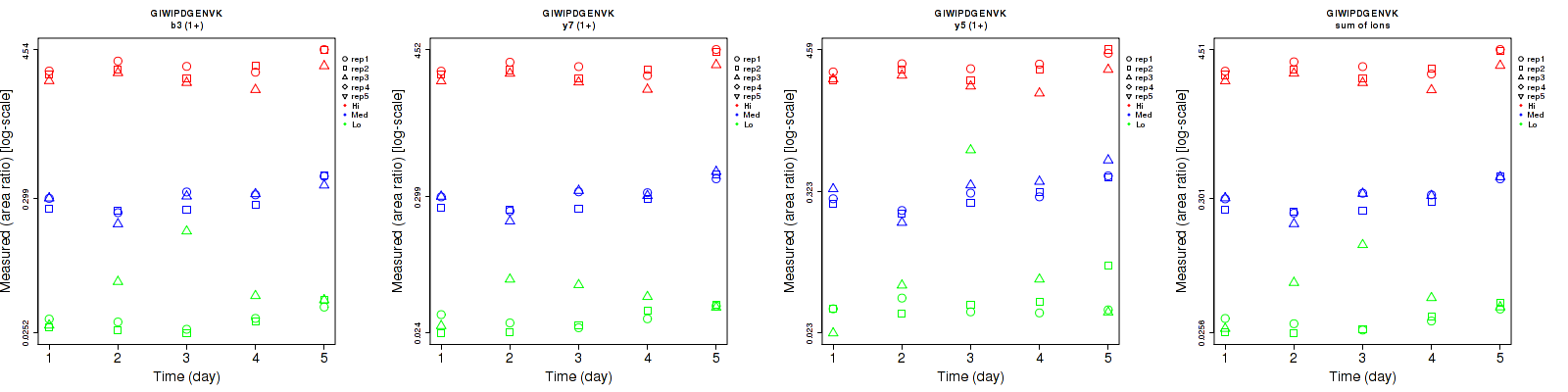
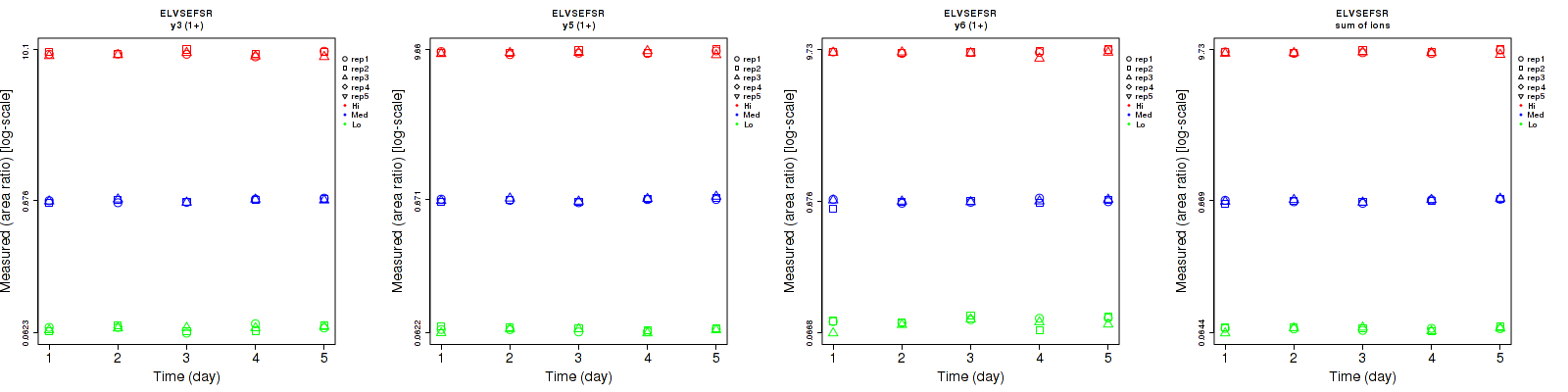
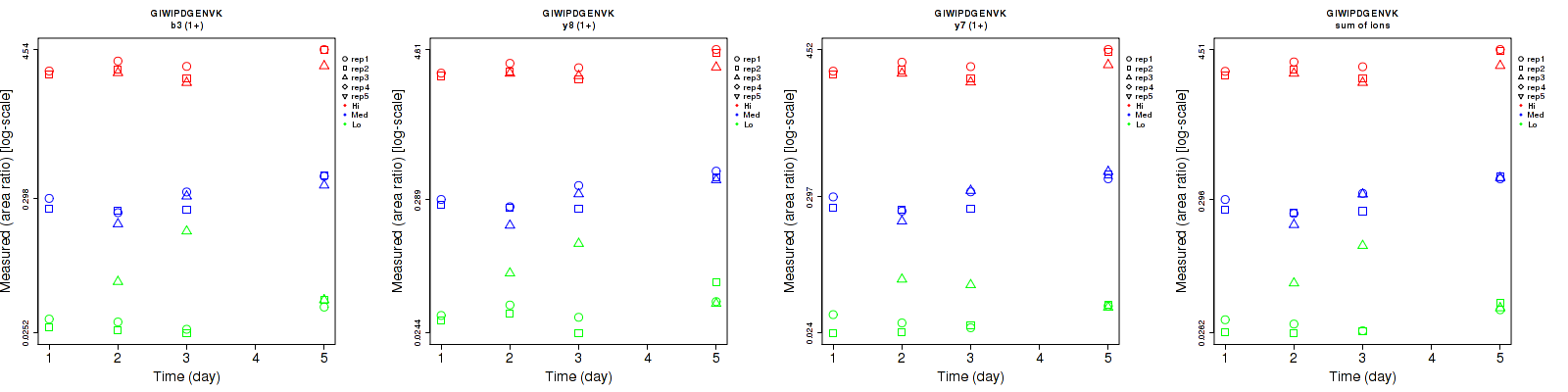
Assay Details for CPTAC-5777 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
GIWIPDGENVK
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
737
Peptide End
747
CPTAC ID
CPTAC-5777
Peptide Molecular Mass
1,226.6295
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Fred Hutchinson Cancer Research Center, H. Lee Moffitt Cancer Center & Research Institute, Broad Institute
Submitting Lab PI
Amanda Paulovich, John Koomen, Steven A. Carr
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
SCIEX 5500 / Thermo Quantiva
Internal Standard
Stable Isotope Labeled Standard
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus K
LC
Dionex Ultimate 3000
Column Packing
C18 PepMap 100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b3 (1+) | 40.7 | 12 | 14.8 | 39.2 | 26.6 | 19 | 56.5 | 29.2 | 24.1 | 15 | 15 | 15 |
| y7 (1+) | 29.1 | 10.5 | 13.7 | 23.9 | 28.1 | 19.3 | 37.7 | 30 | 23.7 | 15 | 15 | 15 |
| y5 (1+) | 57 | 15.1 | 16.4 | 74 | 30.1 | 18.7 | 93.4 | 33.7 | 24.9 | 15 | 15 | 15 |
| sum | 38.2 | 9.8 | 14.1 | 34.4 | 27.4 | 19.1 | 51.4 | 29.1 | 23.7 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b3 (1+) | 40.7 | 12 | 14.8 | 39.2 | 26.6 | 19 | 56.5 | 29.2 | 24.1 | 15 | 15 | 15 |
| y7 (1+) | 29.1 | 10.5 | 13.7 | 23.9 | 28.1 | 19.3 | 37.7 | 30 | 23.7 | 15 | 15 | 15 |
| y5 (1+) | 57 | 15.1 | 16.4 | 74 | 30.1 | 18.7 | 93.4 | 33.7 | 24.9 | 15 | 15 | 15 |
| sum | 38.2 | 9.8 | 14.1 | 34.4 | 27.4 | 19.1 | 51.4 | 29.1 | 23.7 | 15 | 15 | 15 |
Additional Resources and Comments
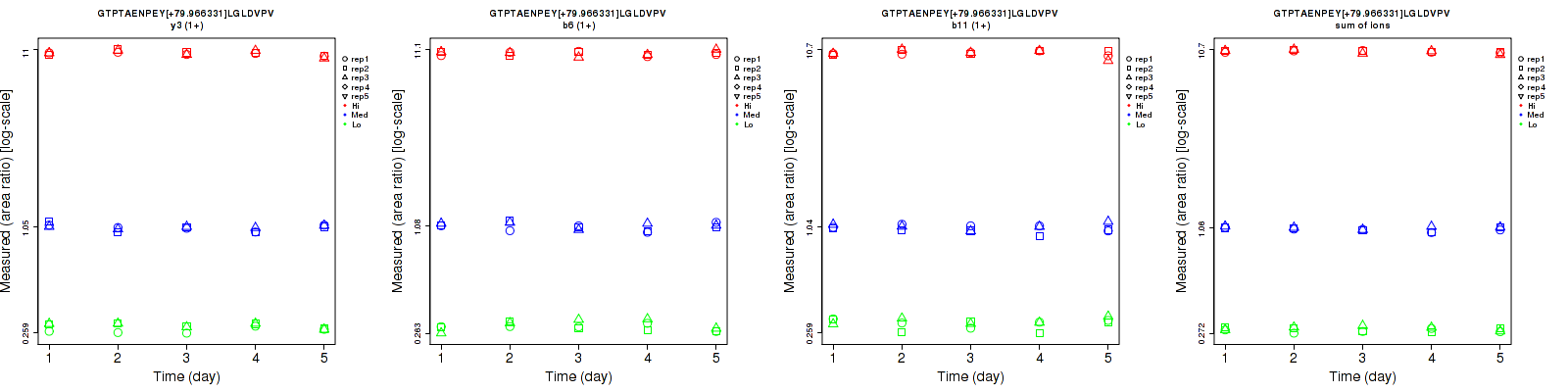
Assay Details for CPTAC-5827 Collapse assay details
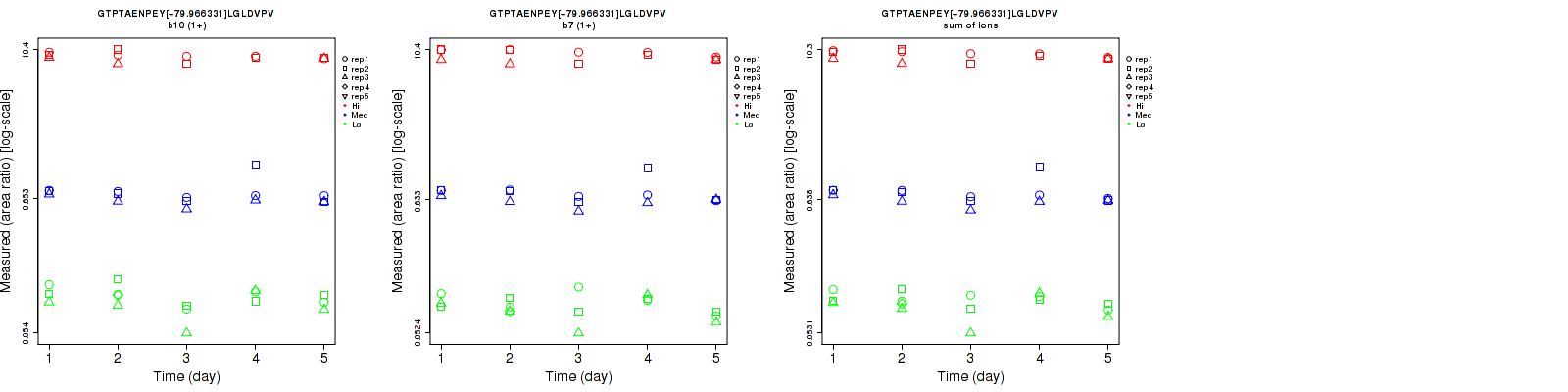
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Modified Sequence
GTPTAENPEY[+79.966331]LGLDVPV
Modification Type
Phospho (Y)
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
1239
Peptide End
1255
CPTAC ID
CPTAC-5827
Peptide Molecular Mass
1,850.8339
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Fred Hutchinson Cancer Research Center, H. Lee Moffitt Cancer Center & Research Institute, Broad Institute
Submitting Lab PI
Amanda Paulovich, John Koomen, Steven A. Carr
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
SCIEX 5500 / Thermo Quantiva
Internal Standard
Stable Isotope Labeled Standard
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Dionex Ultimate 3000
Column Packing
C18 PepMap 100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b10 (1+) | 17.2 | 13.7 | 6.1 | 21.1 | 15.7 | 6.7 | 27.2 | 20.8 | 9.1 | 16 | 15 | 13 |
| b7 (1+) | 16.8 | 13.7 | 9.3 | 20.2 | 15.5 | 7.1 | 26.3 | 20.7 | 11.7 | 16 | 15 | 13 |
| sum | 16.4 | 13.4 | 7.6 | 18.1 | 15.5 | 6.8 | 24.4 | 20.5 | 10.2 | 16 | 15 | 13 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b10 (1+) | 17.2 | 13.7 | 6.1 | 21.1 | 15.7 | 6.7 | 27.2 | 20.8 | 9.1 | 16 | 15 | 13 |
| b7 (1+) | 16.8 | 13.7 | 9.3 | 20.2 | 15.5 | 7.1 | 26.3 | 20.7 | 11.7 | 16 | 15 | 13 |
| sum | 16.4 | 13.4 | 7.6 | 18.1 | 15.5 | 6.8 | 24.4 | 20.5 | 10.2 | 16 | 15 | 13 |
Additional Resources and Comments
Assay Details for non-CPTAC-5385 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
ELVSEFSR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
971
Peptide End
978
CPTAC ID
non-CPTAC-5385
Peptide Molecular Mass
965.4818
Species
Homo sapiens (Human)
Assay Type
Direct MRM
Matrix
Cell Lysate
Submitting Laboratory
Moffitt Cancer Center
Submitting Lab PI
John Koomen
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
RSLCnano and Quantiva
Internal Standard
Stable Isotope Labeled Standard
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus R
LC
RSLCnano
Column Packing
C18 PepMap 100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nl/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y3 (1+) | 3.9 | 1.6 | 3.5 | 4.5 | 2.9 | 3.2 | 6 | 3.3 | 4.7 | 15 | 15 | 15 |
| y4 (1+) | 9.2 | 3 | 3 | 8.7 | 5.7 | 3.6 | 12.7 | 6.4 | 4.7 | 15 | 15 | 15 |
| y5 (1+) | 3.1 | 1.8 | 2.8 | 3 | 3.1 | 2.7 | 4.3 | 3.6 | 3.9 | 15 | 15 | 15 |
| y6 (1+) | 7.2 | 3.7 | 2.4 | 7.5 | 3.8 | 3 | 10.4 | 5.3 | 3.8 | 15 | 15 | 15 |
| sum | 2.9 | 1.5 | 2.1 | 3 | 3 | 2.2 | 4.2 | 3.4 | 3 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y3 (1+) | 3.9 | 1.6 | 3.5 | 4.5 | 2.9 | 3.2 | 6 | 3.3 | 4.7 | 15 | 15 | 15 |
| y4 (1+) | 9.2 | 3 | 3 | 8.7 | 5.7 | 3.6 | 12.7 | 6.4 | 4.7 | 15 | 15 | 15 |
| y5 (1+) | 3.1 | 1.8 | 2.8 | 3 | 3.1 | 2.7 | 4.3 | 3.6 | 3.9 | 15 | 15 | 15 |
| y6 (1+) | 7.2 | 3.7 | 2.4 | 7.5 | 3.8 | 3 | 10.4 | 5.3 | 3.8 | 15 | 15 | 15 |
| sum | 2.9 | 1.5 | 2.1 | 3 | 3 | 2.2 | 4.2 | 3.4 | 3 | 15 | 15 | 15 |
Additional Resources and Comments
Assay Details for non-CPTAC-5547 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
GIWIPDGENVK
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
6
Peptide Start
737
Peptide End
747
CPTAC ID
non-CPTAC-5547
Peptide Molecular Mass
1,226.6295
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
N/A
Matrix
Cell Line
Submitting Laboratory
Moffitt Cancer Center
Submitting Lab PI
John Koomen
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
RSLCnano and Quantiva
Internal Standard
SIS Peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus K
LC
RSLCnano
Column Packing
C18 PepMap100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b3 (1+) | 44.7 | 13 | 12.1 | 39.2 | 32.4 | 19.4 | 59.5 | 34.9 | 22.9 | 11 | 11 | 11 |
| y5 (1+) | 57 | 13.3 | 14.4 | 67.7 | 37.5 | 20.2 | 88.5 | 39.8 | 24.8 | 11 | 11 | 11 |
| y7 (1+) | 32.8 | 12.5 | 11.4 | 23.3 | 34.4 | 19.2 | 40.2 | 36.6 | 22.3 | 11 | 11 | 11 |
| y8 (1+) | 41.1 | 13.8 | 10.3 | 37.1 | 33.2 | 17.1 | 55.4 | 36 | 20 | 11 | 11 | 11 |
| y9 (1+) | 40.5 | 15.6 | 12.7 | 35.8 | 35.5 | 18.7 | 54.1 | 38.8 | 22.6 | 11 | 11 | 11 |
| sum | 42.8 | 11.1 | 11.7 | 34.7 | 33.8 | 19.1 | 55.1 | 35.6 | 22.4 | 11 | 11 | 11 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b3 (1+) | 44.7 | 13 | 12.1 | 39.2 | 32.4 | 19.4 | 59.5 | 34.9 | 22.9 | 11 | 11 | 11 |
| y5 (1+) | 57 | 13.3 | 14.4 | 67.7 | 37.5 | 20.2 | 88.5 | 39.8 | 24.8 | 11 | 11 | 11 |
| y7 (1+) | 32.8 | 12.5 | 11.4 | 23.3 | 34.4 | 19.2 | 40.2 | 36.6 | 22.3 | 11 | 11 | 11 |
| y8 (1+) | 41.1 | 13.8 | 10.3 | 37.1 | 33.2 | 17.1 | 55.4 | 36 | 20 | 11 | 11 | 11 |
| y9 (1+) | 40.5 | 15.6 | 12.7 | 35.8 | 35.5 | 18.7 | 54.1 | 38.8 | 22.6 | 11 | 11 | 11 |
| sum | 42.8 | 11.1 | 11.7 | 34.7 | 33.8 | 19.1 | 55.1 | 35.6 | 22.4 | 11 | 11 | 11 |
Additional Resources and Comments
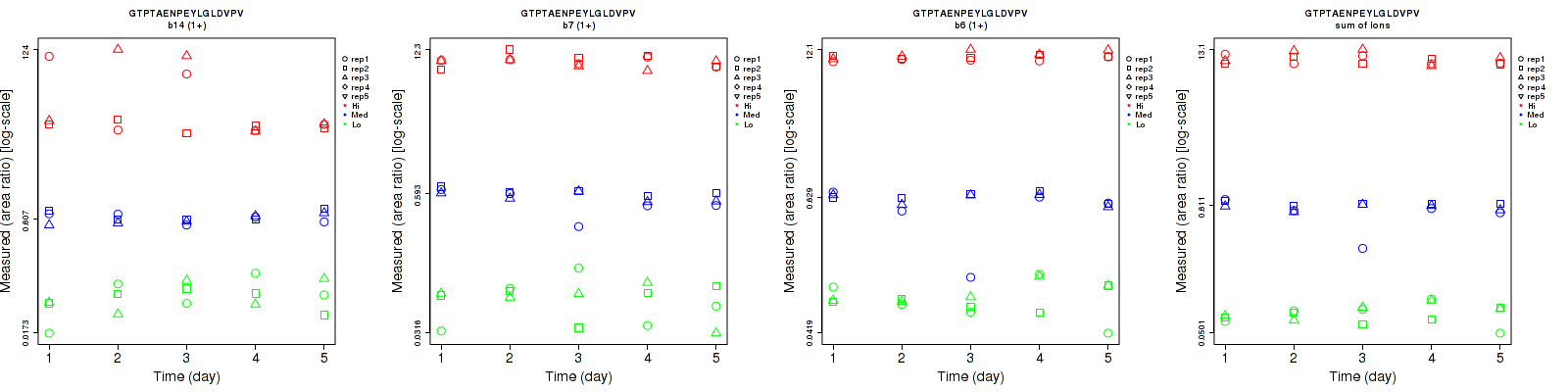
Assay Details for non-CPTAC-5386 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
GTPTAENPEYLGLDVPV
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
1239
Peptide End
1255
CPTAC ID
non-CPTAC-5386
Peptide Molecular Mass
1,770.8676
Species
Homo sapiens (Human)
Assay Type
Direct MRM
Matrix
Cell Lysate
Submitting Laboratory
Moffitt Cancer Center
Submitting Lab PI
John Koomen
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
RSLCnano and Quantiva
Internal Standard
Stable Isotope Labeled Standard
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
RSLCnano
Column Packing
C18 PepMap 100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nl/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b14 (1+) | 45.2 | 14.4 | 70.4 | 45.3 | 16.4 | 76.4 | 64 | 21.8 | 103.9 | 15 | 15 | 15 |
| b7 (1+) | 38.5 | 14.8 | 11 | 39.1 | 15 | 11.4 | 54.9 | 21.1 | 15.8 | 15 | 15 | 15 |
| b6 (1+) | 23.9 | 18.5 | 7 | 29.3 | 22 | 4.9 | 37.8 | 28.7 | 8.5 | 15 | 15 | 15 |
| sum | 15.9 | 13.8 | 9.9 | 17.5 | 13.2 | 9.8 | 23.6 | 19.1 | 13.9 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b14 (1+) | 45.2 | 14.4 | 70.4 | 45.3 | 16.4 | 76.4 | 64 | 21.8 | 103.9 | 15 | 15 | 15 |
| b7 (1+) | 38.5 | 14.8 | 11 | 39.1 | 15 | 11.4 | 54.9 | 21.1 | 15.8 | 15 | 15 | 15 |
| b6 (1+) | 23.9 | 18.5 | 7 | 29.3 | 22 | 4.9 | 37.8 | 28.7 | 8.5 | 15 | 15 | 15 |
| sum | 15.9 | 13.8 | 9.9 | 17.5 | 13.2 | 9.8 | 23.6 | 19.1 | 13.9 | 15 | 15 | 15 |
Additional Resources and Comments
Assay Details for non-CPTAC-5732 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Modified Sequence
GTPTAENPEY[+79.966331]LGLDVPV
Modification Type
Phospho (Y)
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
1239
Peptide End
1255
CPTAC ID
non-CPTAC-5732
Peptide Molecular Mass
1,850.8339
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Moffitt Cancer Center
Submitting Lab PI
John Koomen
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Thermo Quantiva
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N V
LC
Dionex Ultimate 3000 RSLCnano
Column Packing
Pepmap 100
Column Dimensions
75um x 25cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b7 (1+) | 6.9 | 5.8 | 2.7 | 8.2 | 5.7 | 2.7 | 10.7 | 8.1 | 3.8 | 15 | 15 | 15 |
| b6 (1+) | 4.8 | 4.2 | 2.8 | 5.6 | 4.7 | 3 | 7.4 | 6.3 | 4.1 | 15 | 15 | 15 |
| y3 (1+) | 3.9 | 2.4 | 1.7 | 3.4 | 3.8 | 3.2 | 5.2 | 4.5 | 3.6 | 15 | 15 | 15 |
| sum | 3.1 | 1.8 | 1.3 | 2.6 | 2.3 | 1.7 | 4 | 2.9 | 2.1 | 15 | 15 | 15 |
| b11 (1+) | 5.8 | 4.9 | 2.4 | 5.8 | 3.9 | 3.6 | 8.2 | 6.3 | 4.3 | 15 | 15 | 15 |
| b10 (1+) | 6.3 | 5.3 | 3 | 6.1 | 5.2 | 3.5 | 8.8 | 7.4 | 4.6 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b7 (1+) | 6.9 | 5.8 | 2.7 | 8.2 | 5.7 | 2.7 | 10.7 | 8.1 | 3.8 | 15 | 15 | 15 |
| b6 (1+) | 4.8 | 4.2 | 2.8 | 5.6 | 4.7 | 3 | 7.4 | 6.3 | 4.1 | 15 | 15 | 15 |
| y3 (1+) | 3.9 | 2.4 | 1.7 | 3.4 | 3.8 | 3.2 | 5.2 | 4.5 | 3.6 | 15 | 15 | 15 |
| sum | 3.1 | 1.8 | 1.3 | 2.6 | 2.3 | 1.7 | 4 | 2.9 | 2.1 | 15 | 15 | 15 |
| b11 (1+) | 5.8 | 4.9 | 2.4 | 5.8 | 3.9 | 3.6 | 8.2 | 6.3 | 4.3 | 15 | 15 | 15 |
| b10 (1+) | 6.3 | 5.3 | 3 | 6.1 | 5.2 | 3.5 | 8.8 | 7.4 | 4.6 | 15 | 15 | 15 |
Additional Resources and Comments
Assay Details for non-CPTAC-5384 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
LLDIDETEYHADGGK
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
869
Peptide End
883
CPTAC ID
non-CPTAC-5384
Peptide Molecular Mass
1,674.7737
Species
Homo sapiens (Human)
Assay Type
Direct MRM
Matrix
Cell Lysate
Submitting Laboratory
Moffitt Cancer Center
Submitting Lab PI
John Koomen
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
RSLCnano and Quantiva
Internal Standard
Stable Isotope Labeled Standard
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N K
LC
RSLCnano
Column Packing
C18 PepMap 100
Column Dimensions
75 micron ID x 25 cm length
Flow Rate
300 nl/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 11.1 | 6.2 | 2 | 11.3 | 7.5 | 3 | 15.8 | 9.7 | 3.6 | 15 | 15 | 15 |
| y8 (1+) | 17.6 | 6.4 | 3.3 | 18.9 | 8.2 | 3.5 | 25.8 | 10.4 | 4.8 | 15 | 15 | 15 |
| y9 (1+) | 7.5 | 5.1 | 7.1 | 8.2 | 6 | 7.9 | 11.1 | 7.9 | 10.6 | 15 | 15 | 15 |
| y5 (1+) | 10.6 | 5.2 | 4.7 | 12.8 | 4.1 | 4.1 | 16.6 | 6.6 | 6.2 | 15 | 15 | 15 |
| y6 (1+) | 17.4 | 5.1 | 4 | 15 | 4.4 | 4.1 | 23 | 6.7 | 5.7 | 15 | 15 | 15 |
| sum | 6.2 | 4.6 | 3 | 7.1 | 4.1 | 3.2 | 9.4 | 6.2 | 4.4 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 11.1 | 6.2 | 2 | 11.3 | 7.5 | 3 | 15.8 | 9.7 | 3.6 | 15 | 15 | 15 |
| y8 (1+) | 17.6 | 6.4 | 3.3 | 18.9 | 8.2 | 3.5 | 25.8 | 10.4 | 4.8 | 15 | 15 | 15 |
| y9 (1+) | 7.5 | 5.1 | 7.1 | 8.2 | 6 | 7.9 | 11.1 | 7.9 | 10.6 | 15 | 15 | 15 |
| y5 (1+) | 10.6 | 5.2 | 4.7 | 12.8 | 4.1 | 4.1 | 16.6 | 6.6 | 6.2 | 15 | 15 | 15 |
| y6 (1+) | 17.4 | 5.1 | 4 | 15 | 4.4 | 4.1 | 23 | 6.7 | 5.7 | 15 | 15 | 15 |
| sum | 6.2 | 4.6 | 3 | 7.1 | 4.1 | 3.2 | 9.4 | 6.2 | 4.4 | 15 | 15 | 15 |
Additional Resources and Comments
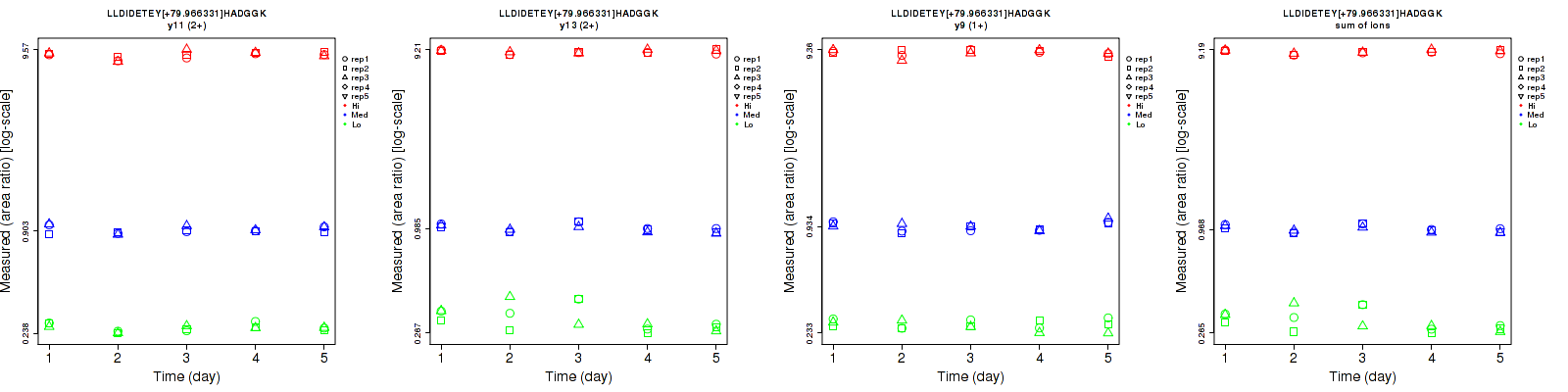
Assay Details for non-CPTAC-5733 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Modified Sequence
LLDIDETEY[+79.966331]HADGGK
Modification Type
Phospho (Y)
Protein - Site of Modification
N/A
Peptide - Site of Modification
3
Peptide Start
869
Peptide End
883
CPTAC ID
non-CPTAC-5733
Peptide Molecular Mass
1,754.7400
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
N/A
Matrix
Cell Lysate
Submitting Laboratory
Moffitt Cancer Center
Submitting Lab PI
John Koomen
Publication
View Details (opens in a new window)
Targeted mass spectrometry-based assays enable multiplex quantification of receptor tyrosine kinase, MAP Kinase, and AKT signaling. Whiteaker JR, Sharma K, Hoffman MA, Kuhn E, Zhao L, Cocco AR, Schoenherr RM, Kennedy JJ, Voytovich U, Lin C, Fang B, Bowers K, Whiteley G, Colantonio S, Bocik W, Roberts R, Hiltke T, Boja E, Rodriguez H, McCormick F, Holderfield M, Carr SA, Koomen JM, Paulovich AG. Cell Reports Methods doi: 10.1016/j.crmeth.2021.100015.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Thermo Quantiva
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N K
LC
Dionex Ultimate 3000 RSLCnano
Column Packing
Pepmap 100
Column Dimensions
75um x 25cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y11 (2+) | 2.8 | 3.5 | 2.7 | 4.7 | 3.9 | 3.9 | 5.5 | 5.2 | 4.7 | 15 | 15 | 15 |
| y13 (2+) | 10.9 | 2.6 | 1.8 | 17 | 5 | 2.2 | 20.2 | 5.6 | 2.8 | 15 | 15 | 15 |
| y7 (1+) | 8.5 | 5.5 | 3.9 | 10 | 6 | 5.7 | 13.1 | 8.1 | 6.9 | 15 | 15 | 15 |
| y9 (1+) | 6.9 | 3.3 | 3 | 6.1 | 5.9 | 4 | 9.2 | 6.8 | 5 | 15 | 15 | 15 |
| sum | 9.3 | 2.3 | 1.3 | 14.3 | 4.4 | 2.2 | 17.1 | 5 | 2.6 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y11 (2+) | 2.8 | 3.5 | 2.7 | 4.7 | 3.9 | 3.9 | 5.5 | 5.2 | 4.7 | 15 | 15 | 15 |
| y13 (2+) | 10.9 | 2.6 | 1.8 | 17 | 5 | 2.2 | 20.2 | 5.6 | 2.8 | 15 | 15 | 15 |
| y7 (1+) | 8.5 | 5.5 | 3.9 | 10 | 6 | 5.7 | 13.1 | 8.1 | 6.9 | 15 | 15 | 15 |
| y9 (1+) | 6.9 | 3.3 | 3 | 6.1 | 5.9 | 4 | 9.2 | 6.8 | 5 | 15 | 15 | 15 |
| sum | 9.3 | 2.3 | 1.3 | 14.3 | 4.4 | 2.2 | 17.1 | 5 | 2.6 | 15 | 15 | 15 |
Additional Resources and Comments
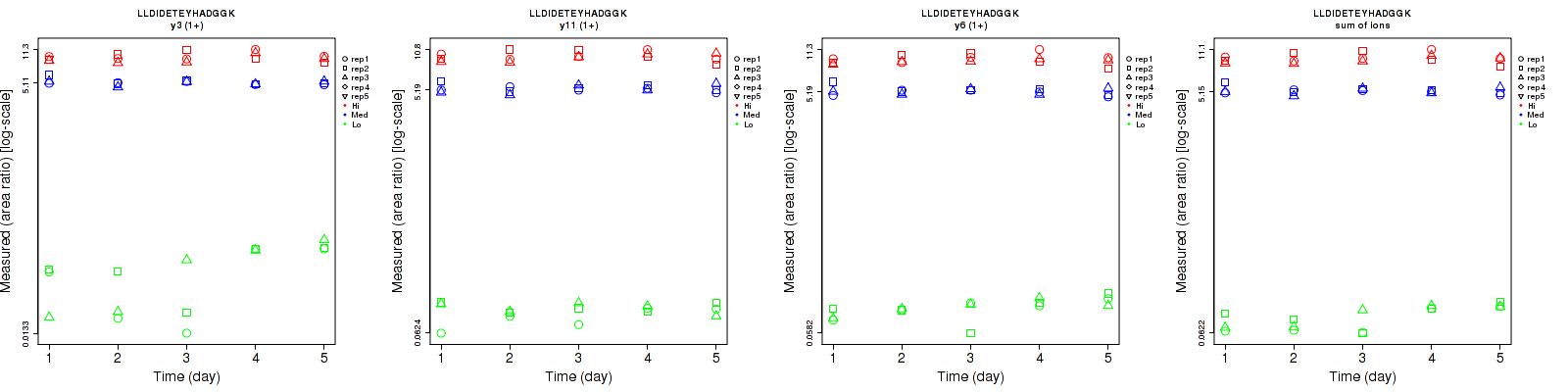
Assay Details for CPTAC-3046 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
LLDIDETEYHADGGK
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
869
Peptide End
883
CPTAC ID
CPTAC-3046
Peptide Molecular Mass
1,674.7737
Species
Homo Sapiens
Assay Type
Direct MRM
Matrix
Tumor Digest
Submitting Laboratory
Washington University in St. Louis
Submitting Lab PI
Reid Townsend
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
ThermoFisher, Q-Exactive
Internal Standard
25 fmol on column
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus K
LC
Easy-nLC 1000 Thermo Scientific
Column Packing
C18
Column Dimensions
75 µm x 50 cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y3 (1+) | 44.1 | 4.3 | 9.8 | 62.8 | 6.2 | 10.4 | 76.7 | 7.5 | 14.3 | 15 | 15 | 15 |
| y11 (1+) | 13.8 | 7.2 | 8.7 | 12.4 | 6.8 | 8.7 | 18.6 | 9.9 | 12.3 | 15 | 15 | 15 |
| y6 (1+) | 12.1 | 6.4 | 8.7 | 17.6 | 6.8 | 8.1 | 21.4 | 9.3 | 11.9 | 15 | 15 | 15 |
| sum | 12.2 | 5.8 | 8.7 | 21.5 | 6.1 | 8.7 | 24.7 | 8.4 | 12.3 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y3 (1+) | 44.1 | 4.3 | 9.8 | 62.8 | 6.2 | 10.4 | 76.7 | 7.5 | 14.3 | 15 | 15 | 15 |
| y11 (1+) | 13.8 | 7.2 | 8.7 | 12.4 | 6.8 | 8.7 | 18.6 | 9.9 | 12.3 | 15 | 15 | 15 |
| y6 (1+) | 12.1 | 6.4 | 8.7 | 17.6 | 6.8 | 8.1 | 21.4 | 9.3 | 11.9 | 15 | 15 | 15 |
| sum | 12.2 | 5.8 | 8.7 | 21.5 | 6.1 | 8.7 | 24.7 | 8.4 | 12.3 | 15 | 15 | 15 |
Additional Resources and Comments
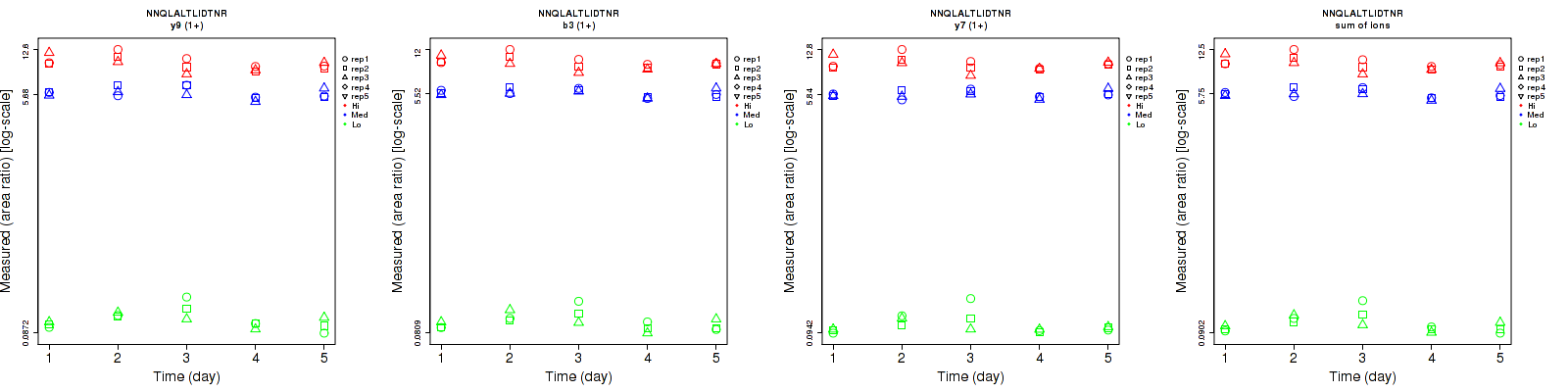
Assay Details for CPTAC-3045 Collapse assay details
Data source: Panorama
Official Gene Symbol
ERBB2
Peptide Sequence
NNQLALTLIDTNR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
176
Peptide End
188
CPTAC ID
CPTAC-3045
Peptide Molecular Mass
1,484.7947
Species
Homo Sapiens
Assay Type
Direct MRM
Matrix
Tumor Digest
Submitting Laboratory
Washington University in St. Louis
Submitting Lab PI
Reid Townsend
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
ThermoFisher, Q-Exactive
Internal Standard
25 fmol on column
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus R
LC
Easy-nLC 1000 Thermo Scientific
Column Packing
C18
Column Dimensions
75 µm x 50 cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y9 (1+) | 9.4 | 6.9 | 9.1 | 16.9 | 9.5 | 12.6 | 19.3 | 11.7 | 15.5 | 15 | 15 | 15 |
| b3 (1+) | 11 | 4.7 | 7.3 | 15.8 | 7 | 10.2 | 19.3 | 8.4 | 12.5 | 15 | 15 | 15 |
| y7 (1+) | 8.8 | 4.7 | 8.1 | 15.2 | 6.4 | 11.2 | 17.6 | 7.9 | 13.8 | 15 | 15 | 15 |
| sum | 9.2 | 5.4 | 8.1 | 15.9 | 7.6 | 11.3 | 18.4 | 9.3 | 13.9 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y9 (1+) | 9.4 | 6.9 | 9.1 | 16.9 | 9.5 | 12.6 | 19.3 | 11.7 | 15.5 | 15 | 15 | 15 |
| b3 (1+) | 11 | 4.7 | 7.3 | 15.8 | 7 | 10.2 | 19.3 | 8.4 | 12.5 | 15 | 15 | 15 |
| y7 (1+) | 8.8 | 4.7 | 8.1 | 15.2 | 6.4 | 11.2 | 17.6 | 7.9 | 13.8 | 15 | 15 | 15 |
| sum | 9.2 | 5.4 | 8.1 | 15.9 | 7.6 | 11.3 | 18.4 | 9.3 | 13.9 | 15 | 15 | 15 |