AOC1, CPTAC-2204 - CPTAC Assay Portal (original) (raw)
Please include the following statement when referencing the CPTAC Assay Portal
We would like to acknowledge the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) Assay Portal (assays.cancer.gov) for developing assays and establishing criteria for the assays described in this publication.
AOC1
Overview Data source: UniProt
| Official Gene Symbol | Other Aliases |
|---|---|
| AOC1 | ABP1, DAO1 |
| Sequence Length (AA) | Molecular Weight (Da) |
|---|---|
| 751 | 85378 |
| Protein Name |
|---|
| Amiloride-sensitive amine oxidase [copper-containing] |
| Sources | |
|---|---|
| UniProt PhosphoSitePlus ® GeneCards | Human Protein Atlas |
Protein Sequence hover to view complete sequence
| 10 | 20 | 30 | 40 | 50 |
|---|---|---|---|---|
| MPALGWAVAA | ILMLQTAMAE | PSPGTLPRKA | GVFSDLSNQE | LKAVHSFLWS |
| 60 | 70 | 80 | 90 | 100 |
| KKELRLQPSS | TTTMAKNTVF | LIEMLLPKKY | HVLRFLDKGE | RHPVREARAV |
| 110 | 120 | 130 | 140 | 150 |
| IFFGDQEHPN | VTEFAVGPLP | GPCYMRALSP | RPGYQSSWAS | RPISTAEYAL |
| 160 | 170 | 180 | 190 | 200 |
| LYHTLQEATK | PLHQFFLNTT | GFSFQDCHDR | CLAFTDVAPR | GVASGQRRSW |
| 210 | 220 | 230 | 240 | 250 |
| LIIQRYVEGY | FLHPTGLELL | VDHGSTDAGH | WAVEQVWYNG | KFYGSPEELA |
| 260 | 270 | 280 | 290 | 300 |
| RKYADGEVDV | VVLEDPLPGG | KGHDSTEEPP | LFSSHKPRGD | FPSPIHVSGP |
| 310 | 320 | 330 | 340 | 350 |
| RLVQPHGPRF | RLEGNAVLYG | GWSFAFRLRS | SSGLQVLNVH | FGGERIAYEV |
| 360 | 370 | 380 | 390 | 400 |
| SVQEAVALYG | GHTPAGMQTK | YLDVGWGLGS | VTHELAPGID | CPETATFLDT |
| 410 | 420 | 430 | 440 | 450 |
| FHYYDADDPV | HYPRALCLFE | MPTGVPLRRH | FNSNFKGGFN | FYAGLKGQVL |
| 460 | 470 | 480 | 490 | 500 |
| VLRTTSTVYN | YDYIWDFIFY | PNGVMEAKMH | ATGYVHATFY | TPEGLRHGTR |
| 510 | 520 | 530 | 540 | 550 |
| LHTHLIGNIH | THLVHYRVDL | DVAGTKNSFQ | TLQMKLENIT | NPWSPRHRVV |
| 560 | 570 | 580 | 590 | 600 |
| QPTLEQTQYS | WERQAAFRFK | RKLPKYLLFT | SPQENPWGHK | RTYRLQIHSM |
| 610 | 620 | 630 | 640 | 650 |
| ADQVLPPGWQ | EEQAITWARY | PLAVTKYRES | ELCSSSIYHQ | NDPWHPPVVF |
| 660 | 670 | 680 | 690 | 700 |
| EQFLHNNENI | ENEDLVAWVT | VGFLHIPHSE | DIPNTATPGN | SVGFLLRPFN |
| 710 | 720 | 730 | 740 | 750 |
| FFPEDPSLAS | RDTVIVWPRD | NGPNYVQRWI | PEDRDCSMPP | PFSYNGTYRP |
| 751 | ||||
| V |
Data source: UniProt
Position of Targeted Peptide Analytes Relative to SNPs, Isoforms, and PTMs
Uniprot Database Entry PhosphoSitePlus ®
Click a point on a node
to view detailed assay information below
All other points link out to UniProt
Phosphorylation Acetylation Ubiquitylation Other
loading

Assay Details for CPTAC-2204 Collapse assay details
Data source: Panorama
Official Gene Symbol
AOC1
Peptide Modified Sequence
LEN[+1.0]ITNPWSPR
Modification Type
Deamidated (N)
Protein - Site of Modification
538
Peptide - Site of Modification
3
Peptide Start
536
Peptide End
546
CPTAC ID
CPTAC-2204
Peptide Molecular Mass
1,326.6568
Species
Homo sapiens (Human)
Assay Type
Direct MRM
Matrix
Human ovarian tissue digest
Submitting Laboratory
Johns Hopkins University
Submitting Lab PI
Daniel Chan, Hui Zhang, Zhen Zhang
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
LCMS-8040
Internal Standard
Stable isotope-labeled peptides
Peptide Standard Purity
Crude (~60%)
Peptide Standard Label Type
13C and 15N at C-terminus R
LC
Nexera X2 UHPLC
Column Packing
C18, 3 µm, 300 Å
Column Dimensions
1.0 mm I.D. x 15 cm
Flow Rate
50 µL/min
Assay Multiplexing Expand assay panel
Johns Hopkins University-G
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

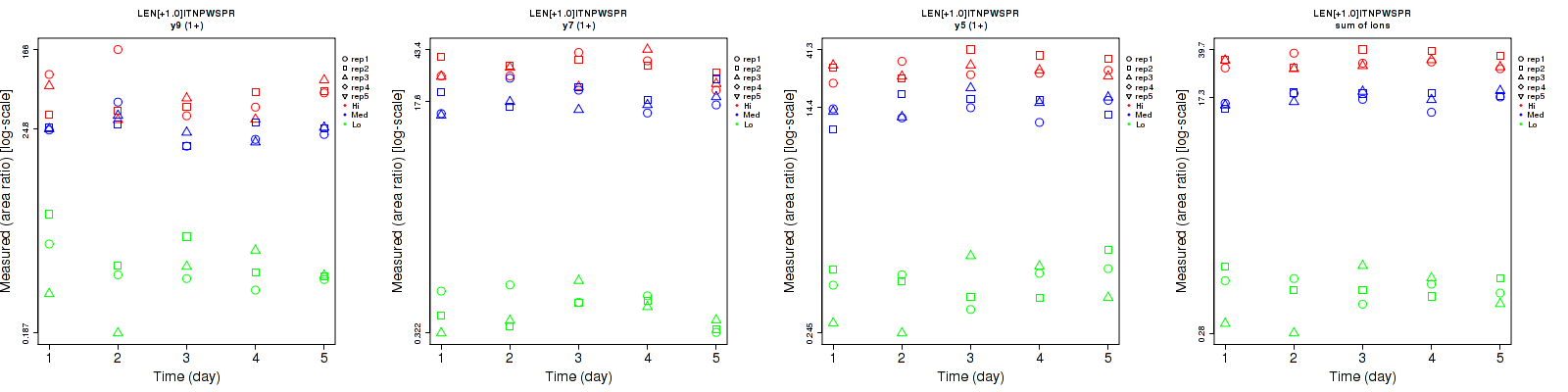
Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y9 (1+) | 49.6 | 17.2 | 42.3 | 59.9 | 30.2 | 45.8 | 77.8 | 34.8 | 62.3 | 15 | 15 | 15 |
| y7 (1+) | 24.4 | 21.2 | 17.6 | 29.3 | 19.6 | 21.3 | 38.1 | 28.9 | 27.6 | 15 | 15 | 15 |
| y5 (1+) | 42.7 | 19.9 | 18.4 | 40.9 | 20.2 | 14.7 | 59.1 | 28.4 | 23.6 | 15 | 15 | 15 |
| sum | 32.2 | 8.6 | 12.5 | 29.6 | 11.5 | 10.2 | 43.7 | 14.4 | 16.1 | 15 | 15 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y9 (1+) | 49.6 | 17.2 | 42.3 | 59.9 | 30.2 | 45.8 | 77.8 | 34.8 | 62.3 | 15 | 15 | 15 |
| y7 (1+) | 24.4 | 21.2 | 17.6 | 29.3 | 19.6 | 21.3 | 38.1 | 28.9 | 27.6 | 15 | 15 | 15 |
| y5 (1+) | 42.7 | 19.9 | 18.4 | 40.9 | 20.2 | 14.7 | 59.1 | 28.4 | 23.6 | 15 | 15 | 15 |
| sum | 32.2 | 8.6 | 12.5 | 29.6 | 11.5 | 10.2 | 43.7 | 14.4 | 16.1 | 15 | 15 | 15 |