KIF5B, CPTAC-397 - CPTAC Assay Portal (original) (raw)
Please include the following statement when referencing the CPTAC Assay Portal
We would like to acknowledge the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) Assay Portal (assays.cancer.gov) for developing assays and establishing criteria for the assays described in this publication.
Protein Sequence hover to view complete sequence
CPTAC-398: View additional LFVQDLATR data
| 10 | 20 | 30 | 40 | 50 |
|---|---|---|---|---|
| MADLAECNIK | VMCRFRPLNE | SEVNRGDKYI | AKFQGEDTVV | IASKPYAFDR |
| 60 | 70 | 80 | 90 | 100 |
| VFQSSTSQEQ | VYNDCAKKIV | KDVLEGYNGT | IFAYGQTSSG | KTHTMEGKLH |
| 110 | 120 | 130 | 140 | 150 |
| DPEGMGIIPR | IVQDIFNYIY | SMDENLEFHI | KVSYFEIYLD | KIRDLLDVSK |
| 160 | 170 | 180 | 190 | 200 |
| TNLSVHEDKN | RVPYVKGCTE | RFVCSPDEVM | DTIDEGKSNR | HVAVTNMNEH |
| 210 | 220 | 230 | 240 | 250 |
| SSRSHSIFLI | NVKQENTQTE | QKLSGKLYLV | DLAGSEKVSK | TGAEGAVLDE |
| 260 | 270 | 280 | 290 | 300 |
| AKNINKSLSA | LGNVISALAE | GSTYVPYRDS | KMTRILQDSL | GGNCRTTIVI |
| 310 | 320 | 330 | 340 | 350 |
| CCSPSSYNES | ETKSTLLFGQ | RAKTIKNTVC | VNVELTAEQW | KKKYEKEKEK |
| 360 | 370 | 380 | 390 | 400 |
| NKILRNTIQW | LENELNRWRN | GETVPIDEQF | DKEKANLEAF | TVDKDITLTN |
| 410 | 420 | 430 | 440 | 450 |
| DKPATAIGVI | GNFTDAERRK | CEEEIAKLYK | QLDDKDEEIN | QQSQLVEKLK |
| 460 | 470 | 480 | 490 | 500 |
| TQMLDQEELL | ASTRRDQDNM | QAELNRLQAE | NDASKEEVKE | VLQALEELAV |
| 510 | 520 | 530 | 540 | 550 |
| NYDQKSQEVE | DKTKEYELLS | DELNQKSATL | ASIDAELQKL | KEMTNHQKKR |
| 560 | 570 | 580 | 590 | 600 |
| AAEMMASLLK | DLAEIGIAVG | NNDVKQPEGT | GMIDEEFTVA | RLYISKMKSE |
| 610 | 620 | 630 | 640 | 650 |
| VKTMVKRCKQ | LESTQTESNK | KMEENEKELA | ACQLRISQHE | AKIKSLTEYL |
| 660 | 670 | 680 | 690 | 700 |
| QNVEQKKRQL | EESVDALSEE | LVQLRAQEKV | HEMEKEHLNK | VQTANEVKQA |
| 710 | 720 | 730 | 740 | 750 |
| VEQQIQSHRE | THQKQISSLR | DEVEAKAKLI | TDLQDQNQKM | MLEQERLRVE |
| 760 | 770 | 780 | 790 | 800 |
| HEKLKATDQE | KSRKLHELTV | MQDRREQARQ | DLKGLEETVA | KELQTLHNLR |
| 810 | 820 | 830 | 840 | 850 |
| KLFVQDLATR | VKKSAEIDSD | DTGGSAAQKQ | KISFLENNLE | QLTKVHKQLV |
| 860 | 870 | 880 | 890 | 900 |
| RDNADLRCEL | PKLEKRLRAT | AERVKALESA | LKEAKENASR | DRKRYQQEVD |
| 910 | 920 | 930 | 940 | 950 |
| RIKEAVRSKN | MARRGHSAQI | AKPIRPGQHP | AASPTHPSAI | RGGGAFVQNS |
| 960 | 963 | |||
| QPVAVRGGGG | KQV |
Data source: UniProt
Position of Targeted Peptide Analytes Relative to SNPs, Isoforms, and PTMs
Uniprot Database Entry PhosphoSitePlus ®
Click a point on a node
to view detailed assay information below
All other points link out to UniProt
Phosphorylation Acetylation Ubiquitylation Other
loading

Assay Details for CPTAC-398 Collapse assay details
Data source: Panorama
Official Gene Symbol
KIF5B
Peptide Sequence
LFVQDLATR
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
802
Peptide End
810
CPTAC ID
CPTAC-398
Peptide Molecular Mass
1,061.5869
Species
Homo sapiens (Human)
Assay Type
Direct MRM
Matrix
cell line lysate pool
Submitting Laboratory
Broad Institute and FHCRC
Submitting Lab PI
Steve Carr
Publication
View Details (opens in a new window)
Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Kennedy JJ, Abbatiello SE, Kim K, Yan P, Whiteaker JR, Lin C, Kim JS, Zhang Y, Wang X, Ivey RG, Zhao L, Min H, Lee Y, Yu MH, Yang EG, Lee C, Wang P, Rodriguez H, Kim Y, Carr SA, Paulovich AG. Nat Methods. 2014 Feb;11(2):149-55. doi: 10.1038/nmeth.2763. Epub 2013 Dec 8. PMID: 24317253
Response curves acquired at Broad Institute. Repeatability data acquired at Fred Hutchinson Cancer Research Center.
Response curves acquired at Broad Institute. Repeatability data acquired at Fred Hutchinson Cancer Research Center.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
5500 QTRAP (ABSCIEX)
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C,15N
LC
nanoLC-Ultra 2D, cHiPLC-nanoflex (Eksigent)
Column Packing
ChromXP C18-CL, 3 um, 120A
Column Dimensions
150 x 0.075 mm
Flow Rate
500 nL / min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

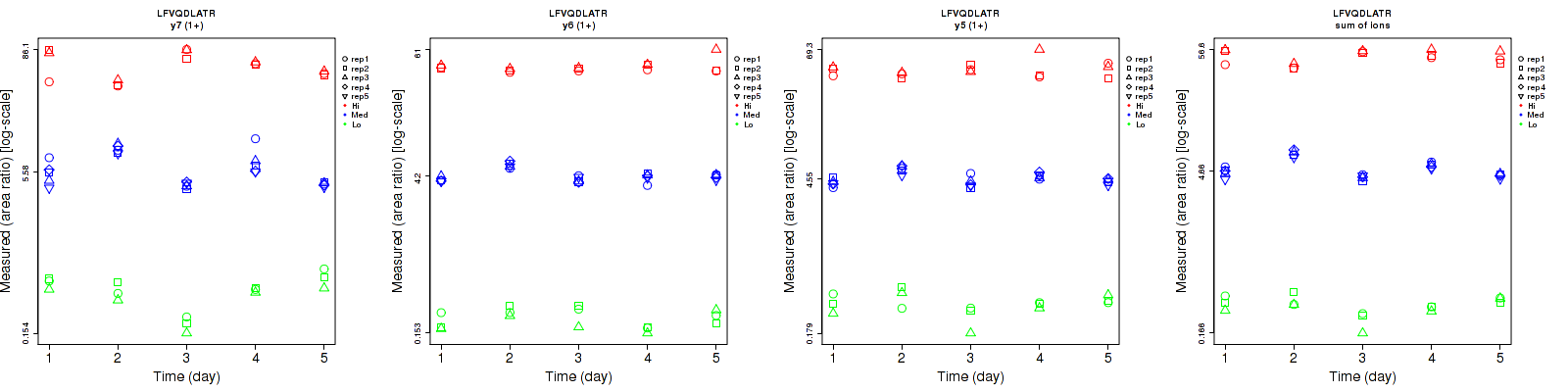
Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 15.4 | 16.1 | 11.8 | 33.7 | 39.1 | 30.4 | 37.1 | 42.3 | 32.6 | 15 | 25 | 15 |
| y6 (1+) | 14.7 | 6.2 | 8.8 | 19.7 | 14.8 | 9.2 | 24.6 | 16 | 12.7 | 15 | 25 | 15 |
| y5 (1+) | 16.5 | 7.9 | 14.9 | 21 | 14.5 | 15.8 | 26.7 | 16.5 | 21.7 | 15 | 25 | 15 |
| sum | 12.2 | 5.9 | 8.9 | 18.6 | 21.3 | 13.1 | 22.2 | 22.1 | 15.8 | 15 | 25 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 15.4 | 16.1 | 11.8 | 33.7 | 39.1 | 30.4 | 37.1 | 42.3 | 32.6 | 15 | 25 | 15 |
| y6 (1+) | 14.7 | 6.2 | 8.8 | 19.7 | 14.8 | 9.2 | 24.6 | 16 | 12.7 | 15 | 25 | 15 |
| y5 (1+) | 16.5 | 7.9 | 14.9 | 21 | 14.5 | 15.8 | 26.7 | 16.5 | 21.7 | 15 | 25 | 15 |
| sum | 12.2 | 5.9 | 8.9 | 18.6 | 21.3 | 13.1 | 22.2 | 22.1 | 15.8 | 15 | 25 | 15 |
Additional Resources and Comments
Assay Details for CPTAC-397 Collapse assay details
Data source: Panorama
Official Gene Symbol
KIF5B
Peptide Sequence
SATLASIDAELQK
Modification Type
unmodified
Protein - Site of Modification
N/A
Peptide - Site of Modification
N/A
Peptide Start
527
Peptide End
539
CPTAC ID
CPTAC-397
Peptide Molecular Mass
1,345.7089
Species
Homo sapiens (Human)
Assay Type
Direct MRM
Matrix
cell line lysate pool
Submitting Laboratory
Broad Institute and FHCRC
Submitting Lab PI
Steve Carr
Publication
View Details (opens in a new window)
Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Kennedy JJ, Abbatiello SE, Kim K, Yan P, Whiteaker JR, Lin C, Kim JS, Zhang Y, Wang X, Ivey RG, Zhao L, Min H, Lee Y, Yu MH, Yang EG, Lee C, Wang P, Rodriguez H, Kim Y, Carr SA, Paulovich AG. Nat Methods. 2014 Feb;11(2):149-55. doi: 10.1038/nmeth.2763. Epub 2013 Dec 8. PMID: 24317253
Response curves acquired at Broad Institute. Repeatability data acquired at Fred Hutchinson Cancer Research Center.
Response curves acquired at Broad Institute. Repeatability data acquired at Fred Hutchinson Cancer Research Center.
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
5500 QTRAP (ABSCIEX)
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C,15N
LC
nanoLC-Ultra 2D, cHiPLC-nanoflex (Eksigent)
Column Packing
ChromXP C18-CL, 3 um, 120A
Column Dimensions
150 x 0.075 mm
Flow Rate
500 nL / min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

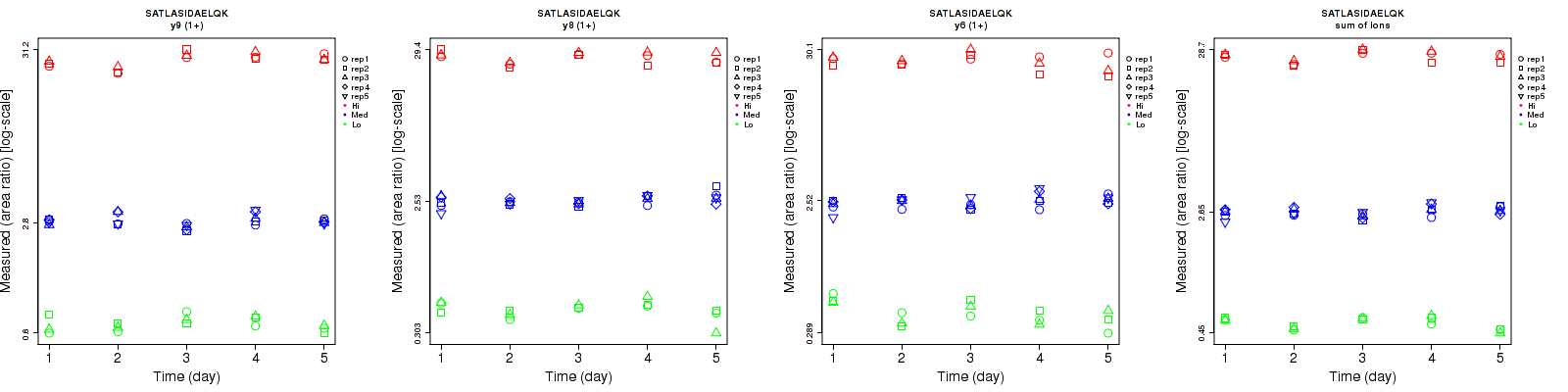
Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y9 (1+) | 8.1 | 5.7 | 4.8 | 10 | 7.6 | 10.1 | 12.9 | 9.5 | 11.2 | 15 | 25 | 15 |
| y8 (1+) | 9.2 | 7.2 | 6.4 | 12.2 | 8.7 | 8.8 | 15.3 | 11.3 | 10.9 | 15 | 25 | 15 |
| y6 (1+) | 12.5 | 9.2 | 10.6 | 19.7 | 10 | 11.1 | 23.3 | 13.6 | 15.3 | 15 | 25 | 15 |
| sum | 3.1 | 6 | 4.8 | 8.7 | 7.3 | 7.8 | 9.2 | 9.4 | 9.2 | 15 | 25 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y9 (1+) | 8.1 | 5.7 | 4.8 | 10 | 7.6 | 10.1 | 12.9 | 9.5 | 11.2 | 15 | 25 | 15 |
| y8 (1+) | 9.2 | 7.2 | 6.4 | 12.2 | 8.7 | 8.8 | 15.3 | 11.3 | 10.9 | 15 | 25 | 15 |
| y6 (1+) | 12.5 | 9.2 | 10.6 | 19.7 | 10 | 11.1 | 23.3 | 13.6 | 15.3 | 15 | 25 | 15 |
| sum | 3.1 | 6 | 4.8 | 8.7 | 7.3 | 7.8 | 9.2 | 9.4 | 9.2 | 15 | 25 | 15 |