TP53, CPTAC-951 - CPTAC Assay Portal (original) (raw)
Please include the following statement when referencing the CPTAC Assay Portal
We would like to acknowledge the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) Assay Portal (assays.cancer.gov) for developing assays and establishing criteria for the assays described in this publication.
Protein Sequence hover to view complete sequence
| 10 | 20 | 30 | 40 | 50 |
|---|---|---|---|---|
| MEEPQSDPSV | EPPLSQETFS | DLWKLLPENN | VLSPLPSQAM | DDLMLSPDDI |
| 60 | 70 | 80 | 90 | 100 |
| EQWFTEDPGP | DEAPRMPEAA | PPVAPAPAAP | TPAAPAPAPS | WPLSSSVPSQ |
| 110 | 120 | 130 | 140 | 150 |
| KTYQGSYGFR | LGFLHSGTAK | SVTCTYSPAL | NKMFCQLAKT | CPVQLWVDST |
| 160 | 170 | 180 | 190 | 200 |
| PPPGTRVRAM | AIYKQSQHMT | EVVRRCPHHE | RCSDSDGLAP | PQHLIRVEGN |
| 210 | 220 | 230 | 240 | 250 |
| LRVEYLDDRN | TFRHSVVVPY | EPPEVGSDCT | TIHYNYMCNS | SCMGGMNRRP |
| 260 | 270 | 280 | 290 | 300 |
| ILTIITLEDS | SGNLLGRNSF | EVRVCACPGR | DRRTEEENLR | KKGEPHHELP |
| 310 | 320 | 330 | 340 | 350 |
| PGSTKRALPN | NTSSSPQPKK | KPLDGEYFTL | QIRGRERFEM | FRELNEALEL |
| 360 | 370 | 380 | 390 | 393 |
| KDAQAGKEPG | GSRAHSSHLK | SKKGQSTSRH | KKLMFKTEGP | DSD |
Data source: UniProt
Position of Targeted Peptide Analytes Relative to SNPs, Isoforms, and PTMs
Uniprot Database Entry PhosphoSitePlus ®
Click a point on a node
to view detailed assay information below
All other points link out to UniProt
Phosphorylation Acetylation Ubiquitylation Other
loading

Assay Details for CPTAC-951 Collapse assay details
Data source: Panorama
Official Gene Symbol
TP53
Peptide Modified Sequence
ALPNNTSSS[+80.0]PQPK
Modification Type
Phospho (ST)
Protein - Site of Modification
315
Peptide - Site of Modification
9
Peptide Start
307
Peptide End
319
CPTAC ID
CPTAC-951
Peptide Molecular Mass
1,419.6395
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
IMAC
Matrix
cell lysate
Submitting Laboratory
Fred Hutchinson Cancer Research Center
Submitting Lab PI
Amanda Paulovich
Publication
View Details (opens in a new window)
Kennedy JJ, Yan P, Zhao L, Ivey RG, Voytovich U, Moore HD, Lin C, Pogosova-Agadjanyan EL, Stirewalt DL, Reding KW, Whiteaker JR, Paulovich AG. Immobilized metal affinity chromatography coupled to multiple reaction monitoring enables reproducible quantification of phospho-signaling. Molecular and Cellular Proteomics. mcp.O115.054940
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
QTrap 5500 (Sciex)
Internal Standard
Peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus K
LC
nanoLC-2D (Eksigent)
Column Packing
ChromXP C18-CL, 3 µm, 120 Å
Column Dimensions
75 µm x 15cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

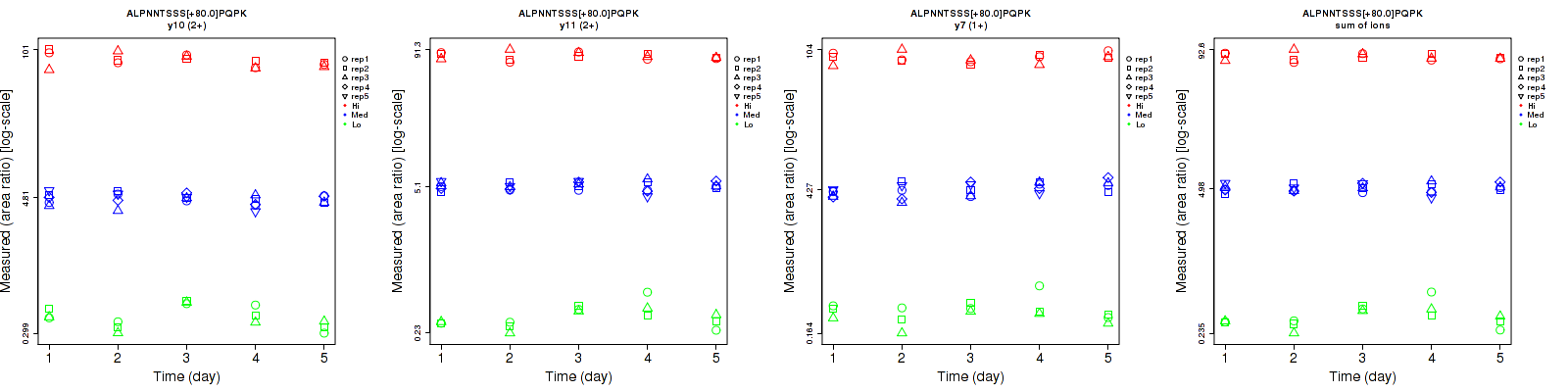
Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b8 (1+) | 63.8 | 71.1 | 45.7 | 60.3 | 78.3 | 101.2 | 87.8 | 105.8 | 111 | 15 | 24 | 15 |
| y10 (2+) | 15.7 | 12.4 | 10 | 33.5 | 14.3 | 14 | 37 | 18.9 | 17.2 | 15 | 24 | 15 |
| y11 (2+) | 12.9 | 11.7 | 6.8 | 24.4 | 11.3 | 7.8 | 27.6 | 16.3 | 10.3 | 15 | 24 | 15 |
| y5 (1+) | 49.6 | 35.7 | 34.7 | 61.6 | 33 | 42.3 | 79.1 | 48.6 | 54.7 | 15 | 24 | 15 |
| y7 (1+) | 22.5 | 13.1 | 10.9 | 23.6 | 14.4 | 11.5 | 32.6 | 19.5 | 15.8 | 15 | 24 | 15 |
| sum | 12.3 | 11.6 | 7.1 | 23.5 | 11.1 | 7.9 | 26.5 | 16.1 | 10.6 | 15 | 24 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b8 (1+) | 63.8 | 71.1 | 45.7 | 60.3 | 78.3 | 101.2 | 87.8 | 105.8 | 111 | 15 | 24 | 15 |
| y10 (2+) | 15.7 | 12.4 | 10 | 33.5 | 14.3 | 14 | 37 | 18.9 | 17.2 | 15 | 24 | 15 |
| y11 (2+) | 12.9 | 11.7 | 6.8 | 24.4 | 11.3 | 7.8 | 27.6 | 16.3 | 10.3 | 15 | 24 | 15 |
| y5 (1+) | 49.6 | 35.7 | 34.7 | 61.6 | 33 | 42.3 | 79.1 | 48.6 | 54.7 | 15 | 24 | 15 |
| y7 (1+) | 22.5 | 13.1 | 10.9 | 23.6 | 14.4 | 11.5 | 32.6 | 19.5 | 15.8 | 15 | 24 | 15 |
| sum | 12.3 | 11.6 | 7.1 | 23.5 | 11.1 | 7.9 | 26.5 | 16.1 | 10.6 | 15 | 24 | 15 |
Additional Resources and Comments
Assay Details for CPTAC-5896 Collapse assay details
Data source: Panorama
Official Gene Symbol
TP53
Peptide Modified Sequence
ALPNNTSSS[+79.966331]PQPK
Modification Type
Phospho (ST)
Protein - Site of Modification
316
Peptide - Site of Modification
9
Peptide Start
307
Peptide End
319
CPTAC ID
CPTAC-5896
Peptide Molecular Mass
1,419.6395
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
IMAC
Matrix
Cell Lysate
Submitting Laboratory
Fred Hutchinson Cancer Research Center
Submitting Lab PI
Amanda Paulovich
Publication
View Details (opens in a new window)
Targeted Mass Spectrometry Enables Quantification of Novel Pharmacodynamic Biomarkers of ATM Kinase Inhibition. Whiteaker JR, Wang T, Zhao L, Schoenherr RM, Kennedy JJ, Voytovich U, Ivey RG, Huang D, Lin C, Colantonio S, Caceres TW, Roberts RR, Knotts JG, Kaczmarczyk JA, Blonder J, Reading JJ, Richardson CW, Hewitt SM, Garcia-Buntley SS, Bocik W, Hiltke T, Rodriguez H, Harrington EA, Barrett JC, Lombardi B, Marco-Casanova P, Pierce AJ, Paulovich AG. Cancers (Basel). 2021 Jul 30;13(15):3843. doi: 10.3390/cancers13153843. PMID: 34359745
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Sciex 5500 QTRAP
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus K
LC
Eksigent 425
Column Packing
Reprosil
Column Dimensions
75um x 15cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

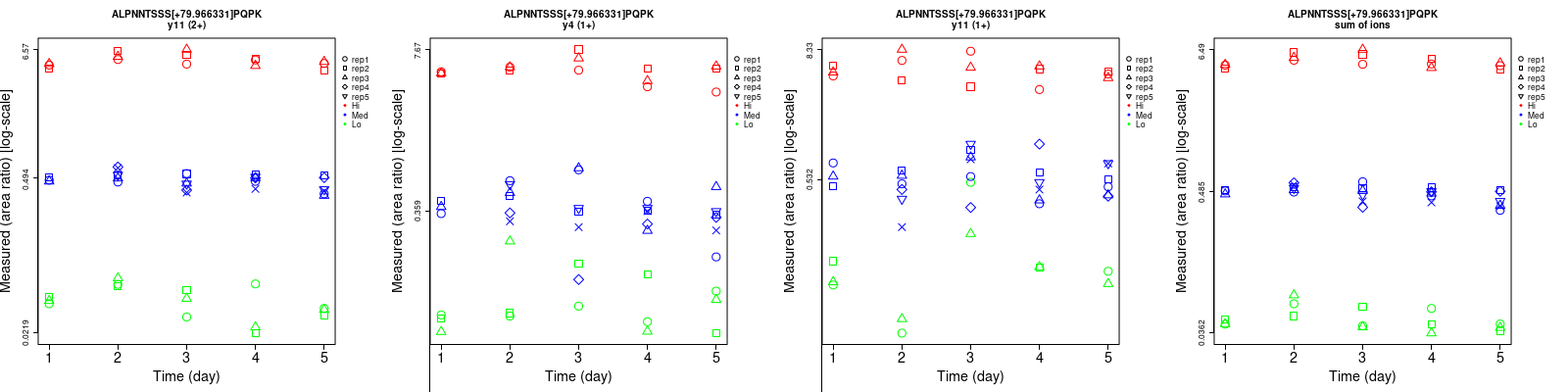
Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y9 (1+) | 141.6 | 41.6 | 26.2 | 173.2 | 54.1 | 29.7 | 223.7 | 68.2 | 39.6 | 15 | 27 | 15 |
| y7 (1+) | 101.9 | 49.6 | 36.9 | 123.4 | 71.9 | 38.6 | 160 | 87.3 | 53.4 | 15 | 27 | 15 |
| y4 (1+) | 61.3 | 33.3 | 13.6 | 65.4 | 32.6 | 18.5 | 89.6 | 46.6 | 23 | 15 | 27 | 15 |
| y11 (2+) | 21.4 | 11.8 | 9.1 | 33.5 | 14.6 | 11.9 | 39.8 | 18.8 | 15 | 15 | 27 | 15 |
| y10 (2+) | 83.4 | 43.1 | 27.8 | 142.6 | 47.8 | 23.7 | 165.2 | 64.4 | 36.5 | 15 | 27 | 15 |
| y11 (1+) | 81.6 | 37.1 | 22.1 | 121.3 | 45.2 | 24.5 | 146.2 | 58.5 | 33 | 15 | 27 | 15 |
| sum | 15.1 | 10.2 | 7.9 | 22.7 | 13 | 10.7 | 27.3 | 16.5 | 13.3 | 15 | 27 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y9 (1+) | 141.6 | 41.6 | 26.2 | 173.2 | 54.1 | 29.7 | 223.7 | 68.2 | 39.6 | 15 | 27 | 15 |
| y7 (1+) | 101.9 | 49.6 | 36.9 | 123.4 | 71.9 | 38.6 | 160 | 87.3 | 53.4 | 15 | 27 | 15 |
| y4 (1+) | 61.3 | 33.3 | 13.6 | 65.4 | 32.6 | 18.5 | 89.6 | 46.6 | 23 | 15 | 27 | 15 |
| y11 (2+) | 21.4 | 11.8 | 9.1 | 33.5 | 14.6 | 11.9 | 39.8 | 18.8 | 15 | 15 | 27 | 15 |
| y10 (2+) | 83.4 | 43.1 | 27.8 | 142.6 | 47.8 | 23.7 | 165.2 | 64.4 | 36.5 | 15 | 27 | 15 |
| y11 (1+) | 81.6 | 37.1 | 22.1 | 121.3 | 45.2 | 24.5 | 146.2 | 58.5 | 33 | 15 | 27 | 15 |
| sum | 15.1 | 10.2 | 7.9 | 22.7 | 13 | 10.7 | 27.3 | 16.5 | 13.3 | 15 | 27 | 15 |
Additional Resources and Comments
Assay Details for CPTAC-5895 Collapse assay details
Data source: Panorama
Official Gene Symbol
TP53
Peptide Modified Sequence
MEEPQSDPSVEPPLS[+79.966331]QETFSDLWK
Modification Type
Phospho (ST)
Protein - Site of Modification
16
Peptide - Site of Modification
15
Peptide Start
1
Peptide End
24
CPTAC ID
CPTAC-5895
Peptide Molecular Mass
2,855.2249
Species
Homo sapiens (Human)
Assay Type
Enrichment MRM
Enrichment Method
IMAC
Matrix
Cell Lysate
Submitting Laboratory
Fred Hutchinson Cancer Research Center
Submitting Lab PI
Amanda Paulovich
Publication
View Details (opens in a new window)
Targeted Mass Spectrometry Enables Quantification of Novel Pharmacodynamic Biomarkers of ATM Kinase Inhibition. Whiteaker JR, Wang T, Zhao L, Schoenherr RM, Kennedy JJ, Voytovich U, Ivey RG, Huang D, Lin C, Colantonio S, Caceres TW, Roberts RR, Knotts JG, Kaczmarczyk JA, Blonder J, Reading JJ, Richardson CW, Hewitt SM, Garcia-Buntley SS, Bocik W, Hiltke T, Rodriguez H, Harrington EA, Barrett JC, Lombardi B, Marco-Casanova P, Pierce AJ, Paulovich AG. Cancers (Basel). 2021 Jul 30;13(15):3843. doi: 10.3390/cancers13153843. PMID: 34359745
Assay Parameters Collapse assay parameters
Data source: Panorama
Instrument
Sciex 5500 QTRAP
Internal Standard
peptide
Peptide Standard Purity
>95%
Peptide Standard Label Type
13C and 15N at C-terminus K
LC
Eksigent 425
Column Packing
Reprosil
Column Dimensions
75um x 15cm
Flow Rate
300 nL/min
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

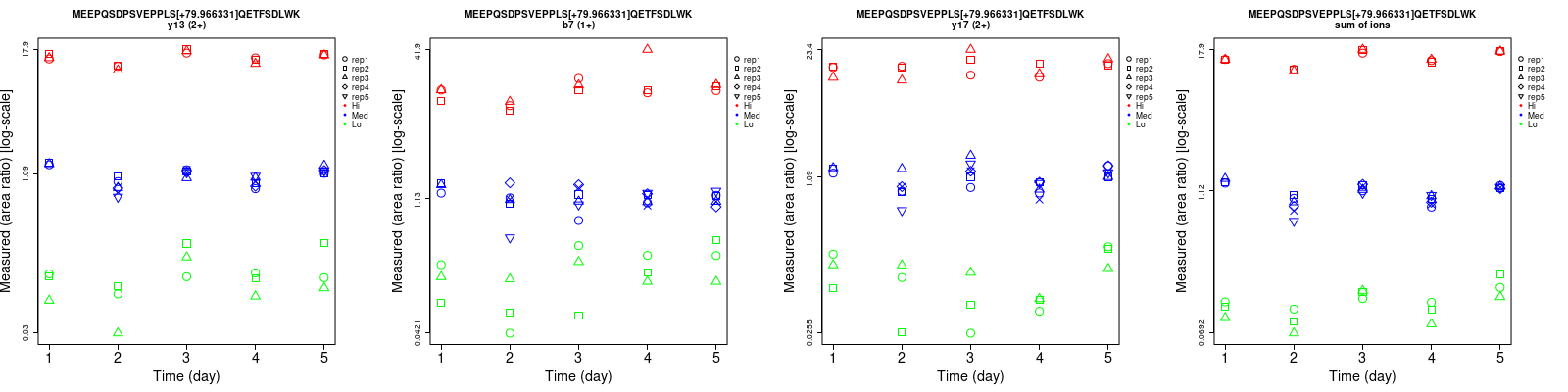
Repeatability
Data source: Panorama
| | Average intra-assay CV(within day CV) | Average inter-assay CV(between day CV) | Total CV | n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b7 (1+) | 51.2 | 20.8 | 22.4 | 53.8 | 24.5 | 34.3 | 74.3 | 32.1 | 41 | 15 | 27 | 15 |
| y12 (2+) | 57 | 24.2 | 19.4 | 63.5 | 30.3 | 20.9 | 85.3 | 38.8 | 28.5 | 15 | 27 | 15 |
| y13 (2+) | 38.8 | 8.7 | 4.9 | 41.9 | 21.1 | 13.7 | 57.1 | 22.8 | 14.5 | 15 | 27 | 15 |
| y17 (2+) | 42.6 | 18.7 | 17.4 | 60.2 | 29 | 18.3 | 73.7 | 34.5 | 25.3 | 15 | 27 | 15 |
| y5 (1+) | 30.3 | 41.5 | 36.3 | 43.2 | 42.7 | 57.6 | 52.8 | 59.5 | 68.1 | 15 | 27 | 15 |
| y10-98 (1+) | 67.6 | 68.3 | 51.3 | 69.4 | 114.1 | 86.9 | 96.9 | 133 | 100.9 | 13 | 26 | 14 |
| b14 (2+) | 59.9 | 98.3 | 31.8 | 99 | 91.8 | 94.4 | 115.7 | 134.5 | 99.6 | 14 | 23 | 14 |
| b11 (1+) | 41 | 21.4 | 9 | 52.7 | 27.6 | 20.4 | 66.8 | 34.9 | 22.3 | 15 | 27 | 15 |
| b10 (1+) | 23.1 | 23.5 | 9.3 | 31.5 | 31.1 | 19.7 | 39.1 | 39 | 21.8 | 15 | 27 | 15 |
| y13-98 (2+) | 43.7 | 26 | 7.4 | 49 | 30.2 | 19.4 | 65.7 | 39.9 | 20.8 | 15 | 27 | 15 |
| sum | 18.1 | 8.1 | 1.8 | 29.9 | 19.9 | 15 | 35 | 21.5 | 15.1 | 15 | 27 | 15 |
| n= | | | | | | | | | |
| ---------------------------------------- | -------------------------------------- | --------------------------------------------------------------------------------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- | ------- | ------- | -------- |
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b7 (1+) | 51.2 | 20.8 | 22.4 | 53.8 | 24.5 | 34.3 | 74.3 | 32.1 | 41 | 15 | 27 | 15 |
| y12 (2+) | 57 | 24.2 | 19.4 | 63.5 | 30.3 | 20.9 | 85.3 | 38.8 | 28.5 | 15 | 27 | 15 |
| y13 (2+) | 38.8 | 8.7 | 4.9 | 41.9 | 21.1 | 13.7 | 57.1 | 22.8 | 14.5 | 15 | 27 | 15 |
| y17 (2+) | 42.6 | 18.7 | 17.4 | 60.2 | 29 | 18.3 | 73.7 | 34.5 | 25.3 | 15 | 27 | 15 |
| y5 (1+) | 30.3 | 41.5 | 36.3 | 43.2 | 42.7 | 57.6 | 52.8 | 59.5 | 68.1 | 15 | 27 | 15 |
| y10-98 (1+) | 67.6 | 68.3 | 51.3 | 69.4 | 114.1 | 86.9 | 96.9 | 133 | 100.9 | 13 | 26 | 14 |
| b14 (2+) | 59.9 | 98.3 | 31.8 | 99 | 91.8 | 94.4 | 115.7 | 134.5 | 99.6 | 14 | 23 | 14 |
| b11 (1+) | 41 | 21.4 | 9 | 52.7 | 27.6 | 20.4 | 66.8 | 34.9 | 22.3 | 15 | 27 | 15 |
| b10 (1+) | 23.1 | 23.5 | 9.3 | 31.5 | 31.1 | 19.7 | 39.1 | 39 | 21.8 | 15 | 27 | 15 |
| y13-98 (2+) | 43.7 | 26 | 7.4 | 49 | 30.2 | 19.4 | 65.7 | 39.9 | 20.8 | 15 | 27 | 15 |
| sum | 18.1 | 8.1 | 1.8 | 29.9 | 19.9 | 15 | 35 | 21.5 | 15.1 | 15 | 27 | 15 |