technology – NIH Director's Blog (original) (raw)
AI Model Takes New Approach to Performing Diagnostic Tasks in Multiple Cancer Types
Posted on October 3rd, 2024 by Dr. Monica M. Bertagnolli

Credit: Donny Bliss/NIH, Adobe Stock
In recent years, medical researchers have been looking for ways to use artificial intelligence (AI) technology for diagnosing cancer. So far, most AI models have been developed to perform specific tasks in cancer diagnosis, such as detecting cancer presence or predicting a tumor’s genetic profile in certain cancer types. But what if an AI system could be more flexible, like a large language model such as ChatGPT, performing a variety of diagnostic tasks across multiple cancer types?
As reported in the journal Nature, researchers have developed an AI system that can perform a wide range of cancer evaluation tasks and outperforms current AI methods in tasks like cancer cell detection and tumor origin identification. It was tested on 19 cancer types, leading the researchers to refer to it as “ChatGPT-like” in its flexibility. According to the research team, whose work is supported in part by NIH, this is also the first AI model based on analyzing slide images to not only accurately predict if a cancer is likely to respond to treatment, but also to validate these predictions across multiple patient groups around the world.
Today, when doctors order a biopsy to find out if cancer is present, those samples are sent to a pathologist, who examines the tissues or cells under a microscope to determine if they are cancerous. The team behind this AI model, led by Kun-Hsing Yu, Harvard Medical School, Boston, recognized that pathologists must analyze a wide variety of disease samples. To make accurate diagnoses in different cancer types, they must take many subtle factors into account.
Most earlier attempts to devise an AI model to analyze tissue samples have depended on training computers to recognize one cancer type at a time. In the new work, the researchers developed a more general-purpose pathology AI system that could analyze a broader range of tissues and sample types. To develop their Clinical Histopathology Imaging Evaluation Foundation (CHIEF) model, the researchers used an AI approach known as self-supervised learning. In this method, a computer is given large volumes of data, in this case 15 million pathology images, to allow it to identify intrinsic patterns and structures. This process allows a computer to “learn” from experience to identify informative features in a vast data set.
The tool was then trained further on more than 60,500 whole-slide images in tissues collected from 19 different parts of the body—such as the lungs, breast, prostate, kidney, brain, and bladder—to bolster the model’s ability to capture similarities and differences among cancer types. This training data was in part comprised of data from The Cancer Genome Atlas (TCGA) program and the Genotype-Tissue Expression (GTEx) Project, both NIH-supported resources. The researchers directed the model to consider both the image as a whole and its finer details, enabling it to interpret the image in a broader context than one region. They then put CHIEF to the test, using another 19,491 whole-slide images from 32 independent slide sets collected from 24 hospitals around the world.
They found that CHIEF worked equally well no matter how the samples were collected (biopsy or surgical excision) and in different clinical settings. In addition to detecting cancers and predicting a cancer’s tissue of origin, CHIEF also predicted with 70% accuracy whether a tissue carried one among dozens of genetic mutations that are commonly seen in cancers. CHIEF showed an ability to predict whether a sample contained mutated copies of 18 genes that oncologists use to make treatment decisions. CHIEF could predict better than earlier models how long a patient was likely to survive following a cancer diagnosis and how aggressively a particular cancer would grow.
This is all good news, but there’s much more work ahead before an AI model like this could be used in the clinic. Next steps for the researchers include training the model on images of tissues from rare cancers, as well as from pre-cancerous and non-cancerous conditions. With continued development and validation, the researchers aim to enable the system to identify cancers most likely to benefit from targeted or experimental therapies in hopes of improving outcomes for more people with cancer in diverse clinical settings around the world.
Reference:
Wang X, et al. A pathology foundation model for cancer diagnosis and prognosis prediction. Nature. DOI: 10.1038/s41586-024-07894-z (2024).
NIH Support: National Institute of General Medical Sciences, National Cancer Institute
Posted In: Health, Science, technology
Tags: AI, artificial intelligence, cancer, cancer diagnosis, computer learning, genetic mutations, genetics, imaging, pathology, technology
Most Detailed 3D Reconstruction of Human Brain Tissue Ever Produced Yields Surprising Insights
Posted on May 30th, 2024 by Dr. Monica M. Bertagnolli
Researchers have developed a detailed 3D reconstruction of a cubic millimeter of brain tissue. Credit: Images in video from Google Research & Lichtman Lab, Harvard University. Renderings by D. Berger, Harvard. Video compiled by Donny Bliss/NIH
The NIH _Brain Research Through Advancing Innovative Neurotechnologies_® (BRAIN) Initiative has expanded scientists’ understanding of the human brain in recent years, offering fascinating insights into the ways that individual cells and complex neural circuits interact dynamically to enable us to think, feel, and act. But neuroscientists still have much more to learn about how our brains are put together at the most fundamental, subcellular level.
As a step in that direction, in a new study supported in part by the NIH BRAIN Initiative and reported in the journal Science, researchers have created the most detailed nanoscale resolution map ever produced of a cubic millimeter of brain tissue, about the size of half a grain of rice.
Despite its small size, this fragment of healthy brain contained about 57,000 cells of various types, 230 millimeters of blood vessels, 150 million neural connections, or synapses, and the protective myelin that insulates neurons. To capture it all in vivid detail, the researchers relied on electron microscopy to amass an impressive 1,400 terabytes of imaging data. For perspective, one terabyte of data is enough to store 100,000 photos on your smartphone.
While there are many more details yet to analyze given the sheer quantity of data, this impressively detailed subcellular map has already revealed multiple brain structures that have never been seen before. This includes a class of triangular neurons in deep brain layers being described for the first time. The map also revealed axons, the long extensions of nerve cells that carry electrical impulses, with as many as 50 synapses and other unusual structures, including axons arranged into extensive spiraling patterns that now warrant further study.
The findings come from a team led by Jeff W. Lichtman, Harvard University, Cambridge, MA, and Viren Jain, Google Research, Mountain View, CA. They recognized that fully understanding the human brain requires knowledge of its most basic construction. While the imaging technologies needed to produce this kind of map were available, there were other barriers, including a limited availability of healthy and high-quality human brain tissue samples for study.
Most biopsies of the brain are done to examine or take out abnormal growths of cells or tissues, making them unsuitable for understanding the normal makeup of the brain. In this case, the researchers were able to obtain a tiny sample from the brain tissue removed and destined for disposal during the normal course of surgery for a patient with epilepsy. The researchers first stained the preserved sample to make the cells easier to trace individually before slicing it into 5,000 thin layers for microscopic imaging.
To put those slices back together into a complete 3D reconstruction, the researchers relied on artificial intelligence (AI) models. Because the dataset is too large for any one group to fully analyze, they’ve made it all freely available to the research community in an online resource. They’ve also provided tools for its further analysis and proofreading.
While there is plenty still left to uncover, the findings offer proof-of-principle that it’s possible to visualize the brain at this very detailed level. This is crucial groundwork for new research now supported by the BRAIN Initiative Connectivity Across Scales (BRAIN CONNECTS) program. BRAIN CONNECTS will develop and scale up tools to produce an equally detailed map of a complete mouse brain, which is about 1,000 times larger than the human brain fragment. The researchers now hope their 3D map and others like it will be put to work to understand both normal and disordered brain function more fully.
Reference:
[1] Shapson-Coe A, et al. A petavoxel fragment of human cerebral cortex reconstructed at nanoscale resolution. Science. DOI: 10.1126/science.adk4858 (2024).
NIH Support: NIH BRAIN Initiative, National Institute of Mental Health
Posted In: News, Science, technology
Tags: 3d reconstruction, brain, BRAIN Initiative, brain tissue, data, imaging, neural circuitry, neuroscience, research tools
Researchers Map Neural Connections Key to Wakefulness in the Human Brain
Posted on May 23rd, 2024 by Dr. Monica M. Bertagnolli

Credit: Donny Bliss/NIH
Human consciousness requires a person to be both awake and aware. While neuroscientists have learned a great deal from research about the underlying brain networks that sustain awareness, surprisingly little has been known about the networks that keep us awake.
Now, an NIH-supported team of researchers has mapped the connectivity of a neural network they suggest is essential for wakefulness, or arousal, in the human brain. According to the researchers, this advance, reported in Science Translational Medicine, is essential for understanding human consciousness. It may also lead to new ways of understanding what happens in the brain when people lose consciousness, with potentially important implications for treating those who have entered a coma or vegetative state.
The team—led by Brian Edlow, Massachusetts General Hospital and Harvard Medical School, Boston, and Hannah Kinney, Boston Children’s Hospital and Harvard Medical School—set out to map the brain network that sustains wakefulness in a manner similar to earlier research that identified the default mode network, which influences awareness. Default networks in the brain are most active when people are at rest rather than focused on a goal-oriented task.
To map what they call the default ascending arousal network, the researchers knew they needed to capture connections deep within human brain areas previously implicated in wakefulness in animal studies. They wanted to use high resolution to uncover fine structural details. Because it isn’t currently possible to capture this in the brain of a living person within a reasonable scan time, the researchers looked to the brains of three organ donors who died without any neurological problems.
The researchers stained different types of brain cells in key areas of the brainstem, hypothalamus, thalamus, and basal forebrain, and took images of the donor brains using a sophisticated form of magnetic resonance imaging (MRI). Their data allowed them to map underlying structures and individual neural connections deep in the brain.
To learn more about how this wakefulness network functions, they next looked to a wealth of functional MRI data from 84 healthy study participants in the NIH-supported Human Connectome Project. Those data revealed functional connections between the arousal network and the previously identified default mode network that is active when people are awake but not attending to their surroundings. Further study revealed a “connectivity hub” between these networks in an area of the midbrain known as the dopaminergic ventral tegmental area, which helps in understanding how arousal and awareness are integrated in human consciousness.
These findings suggest that stimulating this key arousal hub for human consciousness may hold promise for helping people recover from a coma. In fact, the researchers have already launched a clinical trial to see whether stimulating the hub in people in a coma after traumatic brain injury could restore consciousness.
This new guide to brain areas that are essential to wakefulness may ultimately aid understanding of many conditions in which people have altered consciousness, including coma, seizures, and sudden infant death syndrome (SIDS), according to the researchers. And, to enable others to continue studying and uncovering other aspects of human consciousness, the team has made its MRI data, methods, and atlas freely available.
Reference:
[1] Edlow BL, et al. Multimodal MRI reveals brainstem connections that sustain wakefulness in human consciousness. Science Translational Medicine. DOI: 10.1126/scitranslmed.adj4303 (2024).
NIH Support: National Institute of Neurological Disorders and Stroke, National Institute for Biomedical Imaging and Bioengineering, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute on Deafness and Other Communication Disorders, National Institute on Aging, National Institute of Mental Health, NIH BRAIN Initiative
Speeding the Diagnosis of Rare Genetic Disorders with the Help of Artificial Intelligence
Posted on May 16th, 2024 by Dr. Monica M. Bertagnolli

Credit: Donny Bliss/NIH, Qpt/Shutterstock, taka/Adobe Stock
Millions of children around the world are born each year with severe genetic disorders. Many of these are Mendelian disorders, which are rare genetic conditions caused by mutations in a single gene. But pinpointing the specific gene responsible for a disorder to get a clear diagnosis for an individual can be labor-intensive, and reanalysis of undiagnosed cases is also difficult. As a result, only about 30% of people with a rare genetic disorder get a definitive diagnosis, and on average, it takes 6 years from symptom onset to diagnosis.
Progress is needed to get accurate diagnoses to individuals and families more often and faster, and to create more efficient ways to update genetic diagnoses as new discoveries are made. As an important step in this direction, a team funded in part by NIH has developed a new artificial intelligence (AI) system called AI-MARRVEL (AI-Model organism Aggregated Resources for Rare Variant ExpLoration).1
As reported in NEJM AI by a research team led by Pengfei Liu, Hugo Bellen, and Zhandong Liu at the Baylor College of Medicine and the Jan and Dan Duncan Neurological Research Institute at Texas Children’s Hospital in Houston, AI-MARRVEL relies on a machine learning approach. Machine learning involves using vast quantities of data to train computer systems to become increasingly better at recognizing patterns.
The AI-MARRVEL system was trained using a compendium of data called MARRVEL, previously developed by the research team. MARRVEL integrates genetic information from six human databases and seven model organism databases into one site and includes more than 3.5 million known genetic variants from thousands of healthy individuals as well as those with diagnosed cases of genetic disorders. Using what it has learned from that compendium of data, AI-MARRVEL uses a person’s symptoms and protein-coding genome sequences to narrow down the most likely variants responsible for that person’s genetic condition.
To find out how well it works, the researchers compared the results from AI-MARRVEL to other previously published tools for genetic diagnosis based on three different databases containing established molecular diagnoses from a clinical diagnostic laboratory: Baylor Genetics, the NIH-funded Undiagnosed Diseases Network (UDN), and the Deciphering Developmental Disorders (DDD) project. Overall, the researchers found that AI-MARRVEL consistently made accurate diagnoses in twice as many cases as these other tools.
While hundreds of new disease-causing variants are discovered each year, there’s currently no streamlined way to determine which cases should be reanalyzed when previous sequencing and interpretation failed to identify the cause.2 To see how well AI-MARRVEL does at identifying diagnosable cases from pools of unsolved cases, the researchers designed a confidence metric and found the tool achieved a precision rate of 98% and correctly identified 57% of diagnosable cases out of a collection of 871 cases. The researchers also suggest that AI-MARRVEL could help identify short lists of possible gene candidates in even more potentially solvable cases and then send them on to a panel of experts for follow-up review.
There is some early evidence that AI-MARRVEL could also be put to work in making new discoveries that link novel gene variants to diseases for the first time. In fact, the model already correctly identified two recently reported disease genes in a list of top candidates.
These findings suggest a promising path forward where machine learning could one day make diagnostic decisions in a way that’s comparable to experts, only more efficiently. What’s especially exciting is AI-MARRVEL could have the potential for solving rare disease cases, including those that have remained a mystery for years. The hope is that, by combining the power of AI tools together with the latest sequencing data in the years to come, doctors will be able to get faster diagnoses to many more people with rare genetic disorders.
References:
[1] Mao, D, et al. AI-MARRVEL: A Knowledge-Driven Artificial Intelligence for Molecular Diagnostics of Mendelian Disorders. NEJM AI. DOI: 10.1056/AIoa2300009 (2024).
[2] Liu, P, et al. Reanalysis of Clinical Exome Sequencing Data. N Engl J Med. DOI: 10.1056/NEJMc1812033 (2019).
NIH Support: NIH Common Fund, National Human Genome Research Institute, National Institute of Neurological Disorders and Stroke, Eunice Kennedy Shriver National Institute of Child Health and Human Development
Can Bioprinted Skin Substitutes Replace Traditional Grafts for Treating Burn Injuries and Other Serious Skin Wounds?
Posted on October 17th, 2023 by Lawrence Tabak, D.D.S., Ph.D.
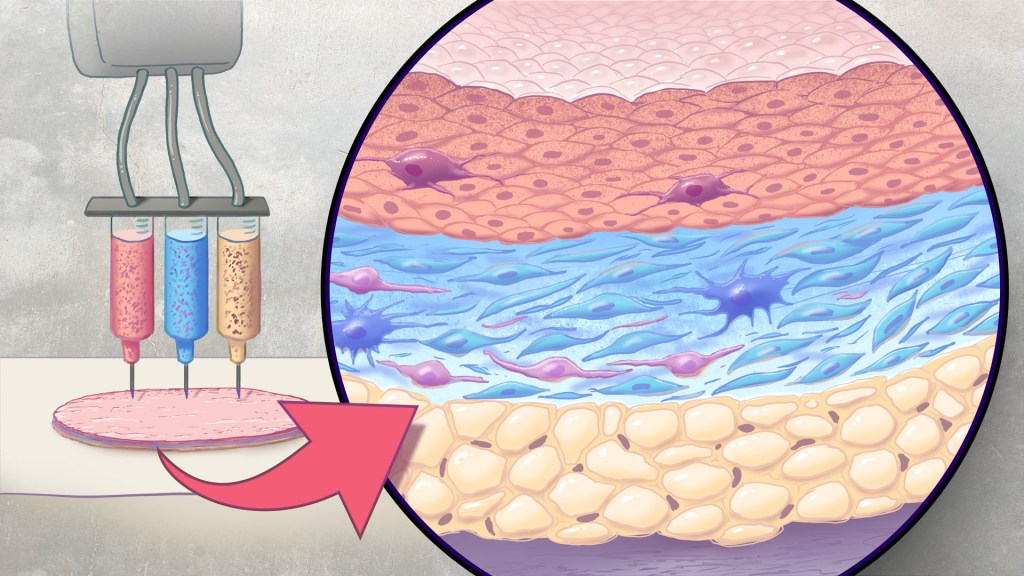
Artificial skin is printed by layering specific cell mixtures to mimic human skin: epidermis (top), dermis (middle) and hypodermis (bottom). Credit: Donny Bliss/NIH
Each year in the U.S., more than 500,000 people receive treatment for burn injuries and other serious skin wounds.1 To close the most severe wounds with less scarring, doctors often must surgically remove skin from one part of a person’s body and use it to patch the injured site. However, this is an intensive process, and some burn patients with extensive skin loss do not have sufficient skin available for grafting. Scientists have been exploring ways to repair these serious skin wounds without skin graft surgery.
An NIH-funded team recently showed that bioprinted skin substitutes may serve as a promising alternative to traditional skin grafts in preclinical studies reported in _Science Translational Medicine._2 The approach involves a portable skin bioprinter system that deposits multiple layers of skin directly into a wound. The recent findings add to evidence that bioprinting technology can successfully regenerate human-like skin to allow healing. While this approach has yet to be tested in people, it confirms that such technologies already can produce skin constructs with the complex structures and multiple cell types present in healthy human skin.
This latest work comes from a team led by Adam Jorgensen and Anthony Atala at Wake Forest School of Medicine’s Wake Forest Institute for Regenerative Medicine, Winston-Salem, NC. Members of the Atala lab and their colleagues had earlier shown it was possible to isolate two major skin cell types found in the skin’s outer (epidermis) and middle (dermis) layers from a small biopsy of healthy skin, expand the number of cells in the lab and then deliver the cells directly into an injury using a specially designed bioprinter.3 Using integrated imaging technology to scan a wound, computer software “prints” cells right into an injury, mimicking two of our skin’s three natural layers.
In the new study, Atala’s team has gone even further to construct skin substitutes that mimic the structure of human skin and that include six primary human skin cell types. They then used their bioprinter to produce skin constructs with all three layers found in healthy human skin: epidermis, dermis, and hypodermis.
To put their skin substitutes to the test, they first transplanted them into mice. Their studies showed that the bioprinted skin encouraged the rapid growth of new blood vessels and had other features of normal-looking, healthy skin. The researchers were able to confirm that their bioprinted skin implants successfully integrated into the animals’ regenerated skin to speed healing.
Studies in a pig model of wound healing added to evidence that such bioprinted implants can successfully repair full-thickness wounds, meaning those that extend through all three layers of skin. The bioprinted skin patches allowed for improved wound healing with less scarring. They also found that the bioprinted grafts encouraged activity in the skin from genes known to play important roles in wound healing.
It’s not yet clear if this approach will work as well in the clinic as it does in the lab. To make it feasible, the researchers note there’s a need for improved approaches to isolating and expanding the needed skin cell types. Nevertheless, these advances come as encouraging evidence that bioprinted skin substitutes could one day offer a promising alternative to traditional skin grafts with the capacity to help even more people with severe burns or other wounds.
References:
[1] Burn Incidence Fact Sheet. American Burn Association
[2] AM Jorgensen, et al. Multicellular bioprinted skin facilitates human-like skin architecture in vivo. Science Translational Medicine DOI: 10.1126/scitranslmed.adf7547 (2023).
[3] M Albanna, et al. In Situ Bioprinting of Autologous Skin Cells Accelerates Wound Healing of Extensive Excisional Full-Thickness Wounds. Scientific Reports DOI: 10.1038/s41598-018-38366-w (2019).
NIH Support: National Institute of Arthritis and Musculoskeletal and Skin Diseases
Posted In: Health, News, Science, technology
Tags: bioprinting, burn, burn treatment, dermis, epidermis, hypodermis, imaging, skin, skin cell, skin graft surgery, skin grafts, wound healing, wounds
Rice-Sized Device Tests Brain Tumor’s Drug Responses During Surgery
Posted on September 19th, 2023 by Lawrence Tabak, D.D.S., Ph.D.

A device implanted into a tumor during surgery delivers tiny doses of up to 20 drugs to determine each treatment’s effects. Credit: Donny Bliss, NIH
Scientists have made remarkable progress in understanding the underlying changes that make cancer grow and have applied this knowledge to develop and guide targeted treatment approaches to vastly improve outcomes for people with many cancer types. And yet treatment progress for people with brain tumors known as gliomas—including the most aggressive glioblastomas—has remained slow. One reason is that doctors lack tests that reliably predict which among many therapeutic options will work best for a given tumor.
Now an NIH-funded team has developed a miniature device with the potential to change this for the approximately 25,000 people diagnosed with brain cancers in the U.S. each year [1]. When implanted into cancerous brain tissue during surgery, the rice-sized drug-releasing device can simultaneously conduct experiments to measure a tumor’s response to more than a dozen drugs or drug combinations. What’s more, a small clinical trial reported in Science Translational Medicine offers the first evidence in people with gliomas that these devices can safely offer unprecedented insight into tumor-specific drug responses [2].
These latest findings come from a Brigham and Women’s Hospital, Boston, team led by Pierpaolo Peruzzi and Oliver Jonas. They recognized that drug-screening studies conducted in cells or tissue samples in the lab too often failed to match what happens in people with gliomas undergoing cancer treatment. Wide variation within individual brain tumors also makes it hard to predict a tumor’s likely response to various treatment options.
It led them to an intriguing idea: Why not test various therapeutic options in each patient’s tumor? To do it, they developed a device, about six millimeters long, that can be inserted into a brain tumor during surgery to deliver tiny doses of up to 20 drugs. Doctors can then remove and examine the drug-exposed cancerous tissue in the laboratory to determine each treatment’s effects. The data can then be used to guide subsequent treatment decisions, according to the researchers.
In the current study, the researchers tested their device on six study volunteers undergoing brain surgery to remove a glioma tumor. For each volunteer, the device was implanted into the tumor and remained in place for about two to three hours while surgeons worked to remove most of the tumor. Next, the device was taken out along with the last piece of a tumor at the end of the surgery for further study of drug responses.
Importantly, none of the study participants experienced any adverse effects from the device. Using the devices, the researchers collected valuable data, including how a tumor’s response changed with varying drug concentrations or how each treatment led to molecular changes in the cancerous cells.
More research is needed to better understand how use of such a device might change treatment and patient outcomes in the longer term. The researchers note that it would take more than a couple of hours to determine how treatments produce less immediate changes, such as immune responses. As such, they’re now conducting a follow-up trial to test a possible two-stage procedure, in which their device is inserted first using minimally invasive surgery 72 hours prior to a planned surgery, allowing longer exposure of tumor tissue to drugs prior to a tumor’s surgical removal.
Many questions remain as they continue to optimize this approach. However, it’s clear that such a device gives new meaning to personalized cancer treatment, with great potential to improve outcomes for people living with hard-to-treat gliomas.
References:
[1] National Cancer Institute Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Brain and Other Nervous System Cancer.
[2] Peruzzi P et al. Intratumoral drug-releasing microdevices allow in situ high-throughput pharmaco phenotyping in patients with gliomas. Science Translational Medicine DOI: 10.1126/scitranslmed.adi0069 (2023).
Links:
Brain Tumors – Patient Version (National Cancer Institute/NIH)
Pierpaolo Peruzzi (Brigham and Women’s Hospital, Boston, MA)
Jonas Lab (Brigham and Women’s Hospital, Boston, MA)
NIH Support: National Cancer Institute, National Institute of Biomedical Imaging and Bioengineering, National Institute of Neurological Disorders and Stroke
Posted In: Health, News, Science, technology
Tags: brain cancer, brain tumor, cancer, cancer treatment, clinical research, clinical study, glioblastoma, glioma, translational medicine, translational science, tumor-specific
New Approach to ‘Liquid Biopsy’ Relies on Repetitive RNA in the Bloodstream
Posted on September 12th, 2023 by Lawrence Tabak, D.D.S., Ph.D.
Researchers have identified segments of noncoding RNA circulating in the blood that are early signs of cancer. Credit: Modified from Adobe Stock/ Andrey Popov; Donny Bliss, NIH
It’s always best to diagnose cancer at an early stage when treatment is most likely to succeed. Unfortunately, far too many cancers are still detected only after cancer cells have escaped from a primary tumor and spread to distant parts of the body. This explains why there’s been so much effort in recent years to develop liquid biopsies, which are tests that can pick up on circulating cancer cells or molecular signs of cancer in blood or other bodily fluids and reliably trace them back to the organ in which a potentially life-threatening tumor is growing.
Earlier methods to develop liquid biopsies for detecting cancers often have relied on the presence of cancer-related proteins and/or DNA in the bloodstream. Now, an NIH-supported research team has encouraging evidence to suggest that this general approach to detecting cancers—including aggressive pancreatic cancers—may work even better by taking advantage of signals from a lesser-known form of genetic material called noncoding RNA.
The findings reported in Nature Biomedical Engineering suggest that the new liquid biopsy approach may aid in the diagnosis of many forms of cancer [1]. The studies show that the sensitivity of the tests varies—a highly sensitive test is one that rarely misses cases of disease. However, they already have evidence that millions of circulating RNA molecules may hold promise for detecting cancers of the liver, esophagus, colon, stomach, and lung.
How does it work? The human genome contains about 3 billion paired DNA letters. Most of those letters are transcribed, or copied, into single-stranded RNA molecules. While RNA is best known for encoding proteins that do the work of the cell, most RNA never gets translated into proteins at all. This noncoding RNA includes repetitive RNA that can be transcribed from millions of repeat elements—patterns of the same few DNA letters occurring multiple times in the genome.
Common approaches to studying RNA don’t analyze repetitive RNA, so its usefulness as a diagnostic tool has been unclear—until recently. Last year, the lab of Daniel Kim at the University of California, Santa Cruz reported [2] that a key genetic mutation that occurs early on in some cancers causes repetitive RNA molecules to be secreted in large quantities from cancer cells, even at the earliest stages of cancer. Non-cancerous cells, by comparison, release much less repetitive RNA.
The findings suggested that liquid biopsy tests that look for this repetitive, noncoding RNA might offer a powerful new way to detect cancers sooner, according to the authors. But first they needed a method capable of measuring it. Due to its oftentimes uncertain functions, the researchers have referred to repetitive, noncoding RNA as “dark matter.”
Using a liquid biopsy platform they developed called COMPLETE-seq, Kim’s team trained computers to detect cancers by looking for patterns in RNA data. The platform enables sequencing and analysis of all protein coding and noncoding RNAs—including any RNA from more than 5 million repeat elements—present in a blood sample. They found that their classifiers worked better when repetitive RNAs were included. The findings lend support to the idea that repetitive, noncoding RNA in the bloodstream is a rich source of information for detecting cancers, which has previously been overlooked.
In a study comparing blood samples from healthy people to those with pancreatic cancer, the COMPLETE-seq technology showed that nearly all people in the study with pancreatic cancer had more repetitive, noncoding RNA in their blood samples compared to healthy people, according to the researchers. They used the COMPLETE-seq test on blood samples from people with other types of cancer as well. For example, their test accurately detected 91% of colorectal cancer samples and 93% of lung cancer samples.
They now plan to look at many more cancer types with samples from additional patients representing a broad range of cancer stages. The goal is to develop a single RNA liquid biopsy test that could detect multiple forms of cancer with a high degree of accuracy and specificity. They note that such a test might also be used to guide treatment decisions and more readily detect a cancer’s recurrence. The hope is that one day a comprehensive liquid biopsy test including coding and noncoding RNA will catch many more cancers sooner, when treatment can be most successful.
References:
[1] RE Reggiardo et al. Profiling of repetitive RNA sequences in the blood plasma of patients with cancer. Nature Biomedical Engineering DOI: 10.1038/s41551-023-01081-7 (2023).
[2] RE Reggiardo et al. Mutant KRAS regulates transposable element RNA and innate immunity via KRAB zinc-finger genes. Cell Reports DOI: 10.1016/j.celrep.2022.111104 (2022).
Links:
Daniel Kim Lab (UC Santa Cruz)
Cancer Screening Overview (National Cancer Institute/NIH)
Early Detection (National Cancer Institute/NIH)
NIH Support: National Cancer Institute, National Heart, Lung, and Blood Institute, National Institute of Diabetes and Digestive and Kidney Diseases
Posted In: Health, News, Science, technology
Tags: cancer, colorectal cancer, Daniel Kim, diagnosing cancer, liquid biopsy, lung cancer, noncoding RNA, pancreatic cancer, repetitive rna, RNA
Mapping Immune Cell “Neighborhoods” in Psoriasis to Understand its Course
Posted on June 13th, 2023 by Lawrence Tabak, D.D.S., Ph.D.

Researchers mapped immune cell “neighborhoods” in the skin of people with psoriasis compared to the healthy skin of people without psoriasis to learn more about the disease course and why it comes with more risk for other health problems. Credit: Donny Bliss, NIH
“Location, location, location.” While most of us know this phrase as a real estate adage, location—specifically that of various cell types—is becoming a key area of investigation in studying human disease. New techniques are enabling scientists to understand where certain cells are with respect to one another and how changes in their activity may affect your overall health.
In one recent example of the power of this approach, NIH-funded researchers [1] used a sophisticated method to map immune cells within human skin to get a more detailed picture of psoriasis, a common, chronic disease in which the immune system becomes overactive leading to skin inflammation. People with psoriasis develop patches of itchy, red, and flaky lesions on their skin, which can be mild to severe. For reasons that aren’t entirely clear, they’re also at higher risk for developing a wide range of other health conditions, including a unique form of arthritis known as psoriatic arthritis, diabetes, mental health issues, heart problems, and more.
The hope is that these newly drawn, precise maps of cellular “neighborhoods” in human skin will help chart the precise course of this disease to understand better the differences between mild and more severe forms. They may also yield important clues as to why people with psoriasis develop other health problems more often than people without psoriasis.
In the new study, a team including Jose Scher and Shruti Naik, NYU Langone, New York, analyzed immune cells within 25 skin samples from 14 volunteers, including those with active psoriasis, those with psoriasis but no active lesions, and people with healthy skin who do not have psoriasis. The researchers relied on a sophisticated approach called spatial transcriptomics [2] to map out what happens at the single-cell level within the samples.
In earlier approaches to single-cell analysis, researchers first would separate cells from the tissue they came from. While they could measure gene activity within those cells at the individual level, they couldn’t put things back together to see how they all fit. With spatial transcriptomics, it’s now possible to molecularly profile single cells to measure their activity in a tissue sample while also mapping their locations with respect to other cells.
The new study led to some intriguing findings. For instance, certain immune cells, specifically B cells, moved to the upper layers of the skin during active disease. That’s notable because prior studies had been unable to capture B cells in the skin adequately, and these cells are thought to play an important role in the disease.
Interestingly, the spatial cellular maps revealed inflammatory regions in both actively inflamed skin and in skin that appeared healthy. This finding highlights the fact that the inflammation that goes with psoriasis can affect the skin, and likely other parts of the body, in ways that aren’t easily observed. In future studies, the researchers want to explore how the presence of psoriasis and its underlying changes in immune cell activity may influence other organs and tissues beneath the skin.
Their fine-scale maps also showed increased gene activity in dozens of molecular pathways that are tied to metabolism and the control of lipid levels. That’s especially interesting because these factors are known to go awry in diabetes and heart conditions, which happen more often in people with psoriasis compared to those without. They also could see in their maps that this altered activity sometimes occurred in clear skin distant from any apparent lesions.
Having discovered such signals with potential consequences for other parts of the body, the researchers report that they’re working to understand how inflammatory immune cells and processes in the skin may lead to more widespread disease processes that affect other parts of the body. They plan to conduct similar studies in larger groups of people with and without active psoriasis lesions and studies following individuals with psoriasis over time. They’ll also explore questions about why people respond differently to the same anti-inflammatory treatment regimens.
To speed the process of discovery, they’ve made their maps and associated data freely available as a resource for the scientific community. About 7.5 million adults in the U.S. and millions more worldwide have psoriasis and associated psoriatic conditions [3]. The hope is that these maps will one day help to steer them toward a healthier future.
References:
[1] Spatial transcriptomics stratifies psoriatic disease severity by emergent cellular ecosystems. Castillo RL, Sidhu I, Dolgalev I, Chu T, Prystupa A, Subudhi I, Yan D, Konieczny P, Hsieh B, Haberman RH, Selvaraj S, Shiomi T, Medina R, Girija PV, Heguy A, Loomis CA, Chiriboga L, Ritchlin C, Garcia-Hernandez ML, Carucci J, Meehan SA, Neimann AL, Gudjonsson JE, Scher JU, Naik S. Sci Immunol. 2023 Jun 8;8(84):eabq7991. doi: 10.1126/sciimmunol.abq7991.
[2] Method of the Year: spatially resolved transcriptomics. Marx V. Nat Methods. 2021 Jan;18(1):9-14. doi: 10.1038/s41592-020-01033-y.
[3] Psoriasis Prevalence in Adults in the United States. Armstrong AW, Mehta MD, Schupp CW, Gondo GC, Bell SJ, Griffiths CEM. JAMA Dermatol. 2021 Aug 1;157(8):940-946. doi: 10.1001/jamadermatol.2021.2007.
Links:
Psoriasis (National Institute of Arthritis and Musculoskeletal and Skin Diseases/NIH)
Jose Scher (NYU Langone Health, New York, NY)
Shruti Naik (NYU Langone Health, New York, NY)
NIH Support: National Cancer Institute, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Center for Advancing Translational Sciences, National Institute of Allergy and Infectious Diseases
Posted In: Health, News, Science, technology
Tags: cellular ecosystems, immune, immune cells, inflammatory skin diseases, psoriasis, single cell, skin, spatial transcriptomics
DNA Barcodes Make for Better Single-Cell Analysis
Posted on April 16th, 2018 by Dr. Francis Collins
Caption: Single-cell analysis helps to reveal subtle, but important, differences among human cells, including many types of brain cells.
Credit: Shutterstock, modified by Ryan M. Mulqueen
Imagine how long it would take to analyze the 37 trillion or so cells that make up the human body if you had to do it by hand, one by one! Still, single-cell analysis is crucial to gaining a comprehensive understanding of our biology. The cell is the unit of life for all organisms, and all cells are certainly not the same. Think about it: even though each cell contains the same DNA, some make up your skin while others build your bones; some of your cells might be super healthy while others could be headed down the road to cancer or Alzheimer’s disease.
So, it’s no surprise that many NIH-funded researchers are hard at work in the rapidly emerging field known as single-cell analysis. In fact, one team recently reported impressive progress in improving the speed and efficiency of a method to analyze certain epigenetic features of individual cells [1]. Epigenetics refers to a multitude of chemical and protein “marks” on a cell’s DNA—patterns that vary among cells and help to determine which genes are switched on or off. That plays a major role in defining cellular identity as a skin cell, liver cell, or pancreatic cancer cell.
The team’s rather simple but ingenious approach relies on attaching a unique combination of two DNA barcodes to each cell prior to analyzing epigenetic marks all across the genome, making it possible for researchers to pool hundreds of cells without losing track of each of them individually. Using this approach, the researchers could profile thousands of individual cells simultaneously for less than 50 cents per cell, a 50- to 100-fold drop in price. The new approach promises to yield important insights into the role of epigenetic factors in our health, from the way neurons in our brains function to whether or not a cancer responds to treatment.
Posted In: Health, News, Science, technology
Tags: barcodes, biotechnology, chromatin, combinatorial indexing, DNA, DNA barcode, DNA methylation, DNA sequencing, epigenetics, neurons, single cell analysis
Wearable Scanner Tracks Brain Activity While Body Moves
Posted on March 27th, 2018 by Dr. Francis Collins
Credit: Wellcome Centre for Human Neuroimaging, University College London.
In recent years, researchers fueled by the BRAIN Initiative and many other NIH-supported efforts have made remarkable progress in mapping the human brain in all its amazing complexity. Now, a powerful new imaging technology promises to further transform our understanding [1]. This wearable scanner, for the first time, enables researchers to track neural activity in people in real-time as they do ordinary things—be it drinking tea, typing on a keyboard, talking to a friend, or even playing paddle ball.
This new so-called magnetoencephalography (MEG) brain scanner, which looks like a futuristic cross between a helmet and a hockey mask, is equipped with specialized “quantum” sensors. When placed directly on the scalp surface, these new MEG scanners can detect weak magnetic fields generated by electrical activity in the brain. While current brain scanners weigh in at nearly 1,000 pounds and require people to come to a special facility and remain absolutely still, the new system weighs less than 2 pounds and is capable of generating 3D images even when a person is making motions.
Posted In: Health, Science, technology, Video
Tags: 3D printing, Autism Spectrum Disorder, brain, brain imaging, BRAIN Initiative, cerebral cortex, diagnostics, functional brain imaging, magnetic field sensor, magnetic fields, magnetoencephalography, MEG brain scanner, Parkinson's disease, primary motor cortex, quantum sensors, QuSpin, wearable devices
