cell biology – NIH Director's Blog (original) (raw)
‘Silent’ Mutations Could Have Unexpectedly Far-Reaching Effects that May Impact Health
Posted on September 26th, 2024 by Dr. Monica M. Bertagnolli

Credit: Donny Bliss/NIH
Genetic mutations affect nearly all human diseases. Some genetic disorders such as cystic fibrosis are caused by mutations in a single gene that a person inherits from their parents. Other diseases can be caused by changes in multiple genes or from a combination of gene mutations and environmental factors. We still have a lot to learn about the complex ways that variations in our genes affect health and disease.
Researchers investigating genetic disorders have primarily studied mutations that cause our cells to alter the makeup of proteins, like the most common mutations that cause cystic fibrosis. Less research has been done on alterations called synonymous mutations, which have been called “silent” because they don’t alter the makeup of proteins, leading scientists to long assume that these kinds of mutations don’t produce any noticeable differences in our biology or health. However, recent research has shown that synonymous mutations can lead to significant changes in a cell’s ability to survive and grow. A new NIH-supported study reported in Proceedings of the National Academy of Sciences sheds additional light on the impact of synonymous mutations and their effect on the way proteins are made.
The researchers behind this study, at the University of Notre Dame in Notre Dame, IN, wanted to understand how synonymous mutations may affect how much protein is made and whether proteins are folded correctly in cells. Misfolded proteins are known to play roles in numerous diseases, including cystic fibrosis, Alzheimer’s disease, and some cancers. The study team, led by Patricia L. Clark, who received an NIH Director’s Pioneer Award in 2021 for this work, has shown that synonymous mutations in a particular gene in Escherichia coli (E. coli) bacteria can alter how the encoded protein folds as it is being made, by altering the rate at which cells produce each copy of the protein. The new research goes a step further and shows that silent mutations in one gene can affect the amount of protein produced from a separate, neighboring gene.
Clark and colleagues, including first author Anabel Rodriguez, created nine synonymous versions of the E. coli chloramphenicol acetyltransferase (cat) gene. This gene encodes a protein enzyme that influences the bacterium’s sensitivity to a particular antibiotic. The researchers found that four of the nine synonymous cat gene sequences significantly affected the number of protein enzymes the E. coli produced from this gene. Unlike in prior work from Clark’s lab, the reason for those differences didn’t stem from changes in protein folding. The differences began one step earlier, in the synthesis of RNA copies, or transcripts, from the cat gene’s DNA. An RNA transcript carries the instructions cells use to produce proteins.
How did it happen? The researchers found that some of the synonymous mutations created new sites on the cat gene where the enzyme that synthesizes RNA could bind. As a result, E. coli cells started making RNA transcripts from part of the cat gene and the entire neighboring gene. These new RNA transcripts led those bacterial cells to make more of the protein encoded by the neighboring gene. In short, these findings unexpectedly showed that synonymous mutations in one gene can alter the production of the protein from other genes.
This discovery in E. coli may have important implications for understanding the bacteria’s biology and evolution. Clark’s team continues to study this system to learn more. Their findings may also prove to have broader implications for biology, including for some genetic disorders. It’s an area that warrants more study and attention, to better understand the roles that synonymous mutations may be playing in genes and their effects on human health.
Reference:
Rodriguez A, et al. Synonymous codon substitutions modulate transcription and translation of a divergent upstream gene by modulating antisense RNA production. Proceedings of the National Academy of Sciences. DOI: 10.1073/pnas.2405510121 (2024).
NIH Support: National Institute of General Medical Sciences
Understanding Causes of Devastating Neurodegenerative Condition Affecting Children
Posted on July 18th, 2023 by Lawrence Tabak, D.D.S., Ph.D.

Researchers studied the lack of functional CLN3 protein, which underlies Batten disease. They found lack of the protein leads to a breakdown of the M6PR receptor (green) in the lysosomes and subsequent disruption of needed lysosomal enzymes and the formation of normal lysosomes. Credit: Donny Bliss, NIH
A common theme among parents and family members caring for a child with the rare Batten disease is “love, hope, cure.” While inspiring levels of love and hope are found among these amazing families, a cure has been more elusive. One reason is rooted in the need for more basic research. Although researchers have identified an altered gene underlying Batten disease, they’ve had difficulty pinpointing where and how the gene’s abnormal protein product malfunctions, especially in cells within the nervous system.
Now, this investment in more basic research has paid off. In a paper just published in the journal Nature Communications, an international research team pinpointed where and how a key cellular process breaks down in the nervous system to cause Batten disease, sometimes referred to as CLN3 disease [1]. While there’s still a long way to go in learning exactly how to overcome the cellular malfunction, the findings mark an important step forward toward developing targeted treatments for Batten disease and progress in the quest for a cure.
The research also offers yet another excellent example of how studying rare diseases helps to advance our fundamental understanding of human biology. It shows that helping those touched by Batten disease can shed a brighter light on basic cellular processes that drive other diseases, rare and common.
Batten disease affects about 14,000 people worldwide [2]. For those with the juvenile form of this inherited disease of the nervous system, parents may first notice their seemingly healthy child has difficulty saying words, sudden problems with vision or movement, and changes in behavior. Tragically for parents, with no approved treatments to reverse these symptoms, the disease will worsen, leading to severe vision loss, frequent seizures, and impaired motor skills. The disease can be fatal as early as late childhood or the teenage years.
Batten disease also goes by the more technical name of juvenile neuronal ceroid lipofuscinosis. Using this technical name, it represents one of the more than 70 medically recognized lysosomal storage disorders.
These disorders share a breakdown in the ability of membrane-bound cellular components, known as lysosomes, to degrade the molecular waste products of normal cell biology. As a result, all this undegraded material builds up and eventually kills affected cells. In people with Batten disease, the lost and damaged cells cause progressive dysfunction within the nervous system.
Researchers have known for a while that the most common cause of this breakdown in people with Batten disease is the inheritance of two defective copies of a gene called CLN3. As mentioned above, what’s been missing is a more detailed understanding of what exactly a working copy of the CLN3 gene does and how its loss leads to the changes seen in those with this condition.
Hoping to solve this puzzle was an NIH-supported basic research team led by Alessia Calcagni and Andrea Ballabio, Baylor College of Medicine and Texas Children’s Hospital, Houston, and Telethon Institute of Genetics and Medicine, Naples, Italy.
As described in their latest paper, the researchers first generated an antibody that allowed them to visualize where in cells the protein encoded by CLN3 is found. Their studies unexpectedly showed that this protein has a role outside, not inside, the cell’s estimated 50-to-1,000 lysosomes. Before reaching the lysosomes, the protein first moves through another cellular component called the Golgi body, where many proteins are packaged.
They then identified all the other proteins that interact with the CLN3 protein in the Golgi body and elsewhere in the cell. Their data showed that CLN3 interacts with proteins known for transporting other proteins within the cell and forming new lysosomes.
That gave them a valuable clue: the CLN3 gene must be a player in these fundamentally important cellular processes of protein transport and lysosome formation. Among the proteins CLN3 interacts with in the Golgi body is a particular receptor called M6PR. The receptor known for its role in recognizing lysosomal enzymes and delivering them to the lysosomes, where they go to work inside these bubble-like structures degrading cellular waste products.
The researchers found that loss of CLN3 led this important M6PR receptor to be broken down within lysosomes. The breakdown, in turn, altered the normal shape of new lysosomes, and that limits their functionality. The researchers also showed that restoring CLN3 in cells that lacked this gene also restored the production of more functional lysosomes and lysosomal enzymes.
Overall, the findings point to a major role for CLN3 in the formation of lysosomes and their ability to function. Importantly, the findings also offer clues for understanding the mechanisms that underlie other forms of lysosomal storage disease, which collectively affect as many as one in every 40,000 people [3]. The work also may have broader implications for common neurodegenerative diseases, such as Parkinson’s and Alzheimer’s disease.
Most of all, this paper demonstrates the power of basic research to define needed molecular targets. It shows how these fundamental studies are helping families affected by Batten disease and supporting their love, hope, and quest for a cure.
References:
[1] Loss of the batten disease protein CLN3 leads to mis-trafficking of M6PR and defective autophagic-lysosomal reformation. Calcagni’ A, Staiano L, Zampelli N, Minopoli N, Herz NJ, Cullen PJ, Parenti G, De Matteis MA, Grumati P, Ballabio A, et al. Nat Commun. 2023 Jul 3;14(1):3911. doi: 10.1038/s41467-023-39643-7. PMID: 37400440; PMCID: PMC10317969.
[2] Batten Disease. Boston Children’s Hospital.
[3] Lysosomal storage diseases. Cleveland Clinic fact sheet, June 27, 2022.
Links:
Batten Disease (National Institute of Neurological Disorders and Stroke/NIH)
Rare Diseases (NIH)
Alessia Calcagni (Baylor College of Medicine, Houston, TX)
Andrea Ballabio (Telethon Institute of Genetics and Medicine, Naples, Italy)
NIH Support: National Institute of Neurological Disorders and Stroke; National Cancer Institute; National Center for Advancing Translational Sciences
Posted In: News
Tags: basic research, Batten disease, cell biology, CLN disease, CLN3 gene, Golgi body, lysosomal storage disease, lysosome, M6PR, nervous system, neurodegenerative disorders, neuronal ceroid lipofuscinosis, rare diseaes
Basic Researchers Discover Possible Target for Treating Brain Cancer
Posted on May 23rd, 2023 by Lawrence Tabak, D.D.S., Ph.D.
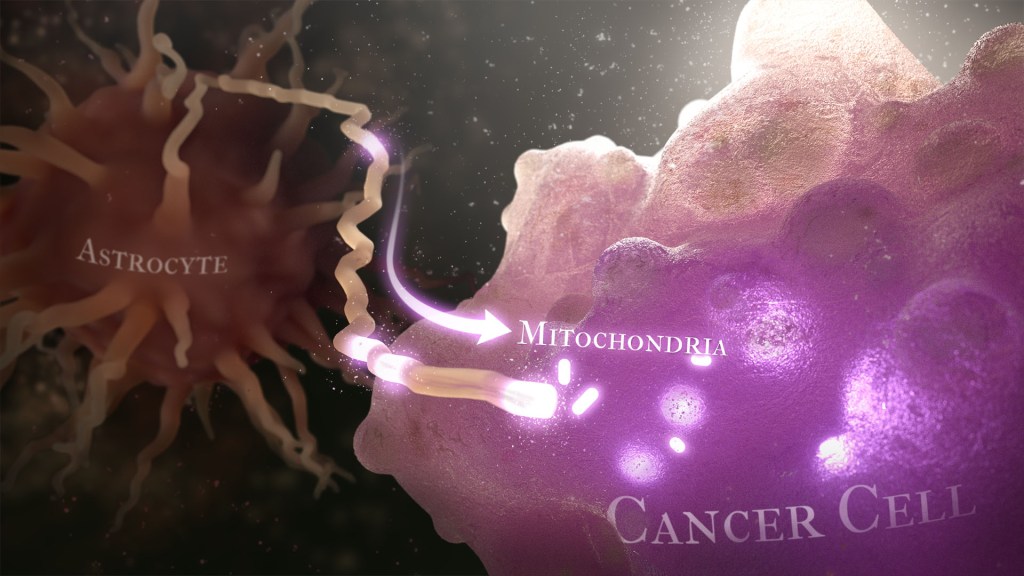
Caption: Illustration of cancer cell (bottom right) stealing mitochondria (white ovals) from a healthy astrocyte cell (left). Credit: Donny Bliss/NIH
Over the years, cancer researchers have uncovered many of the tricks that tumors use to fuel their growth and evade detection by the body’s immune system. More tricks await discovery, and finding them will be key in learning to target the right treatments to the right cancers.
Recently, a team of researchers demonstrated in lab studies a surprising trick pulled off by cells from a common form of brain cancer called glioblastoma. The researchers found that glioblastoma cells steal mitochondria, the power plants of our cells, from other cells in the central nervous system [1].
Why would cancer cells do this? How do they pull it off? The researchers don’t have all the answers yet. But glioblastoma arises from abnormal astrocytes, a particular type of the glial cell, a common cell in the brain and spinal cord. It seems from their initial work that stealing mitochondria from neighboring normal cells help these transformed glioblastoma cells to ramp up their growth. This trick might also help to explain why glioblastoma is one of the most aggressive forms of primary brain cancer, with limited treatment options.
In the new study, published in the journal Nature Cancer, a team co-led by Justin Lathia, Lerner Research Institute, Cleveland Clinic, OH, and Hrvoje Miletic, University of Bergen, Norway, had noticed some earlier studies suggesting that glioblastoma cells might steal mitochondria. They wanted to take a closer look.
This very notion highlights an emerging and much more dynamic view of mitochondria. Scientists used to think that mitochondria—which can number in the thousands within a single cell—generally just stayed put. But recent research has established that mitochondria can move around within a cell. They sometimes also get passed from one cell to another.
It also turns out that the intercellular movement of mitochondria has many implications for health. For instance, the transfer of mitochondria helps to rescue damaged tissues in the central nervous system, heart, and respiratory system. But, in other circumstances, this process may possibly come to the rescue of cancer cells.
While Lathia, Miletic, and team knew that mitochondrial transfer was possible, they didn’t know how relevant or dangerous it might be in brain cancers. To find out, they studied mice implanted with glioblastoma tumors from other mice or people with glioblastoma. This mouse model also had been modified to allow the researchers to trace the movement of mitochondria.
Their studies show that healthy cells often transfer some of their mitochondria to glioblastoma cells. They also determined that those mitochondria often came from healthy astrocytes, a process that had been seen before in the recovery from a stroke.
But the transfer process isn’t easy. It requires that a cell expend a lot of energy to form actin filaments that contract to pull the mitochondria along. They also found that the process depends on growth-associated protein 43 (GAP43), suggesting that future treatments aimed at this protein might help to thwart the process.
Their studies also show that, after acquiring extra mitochondria, glioblastoma cells shift into higher gear. The cancerous cells begin burning more energy as their metabolic pathways show increased activity. These changes allow for more rapid and aggressive growth. Overall, the findings show that this interaction between healthy and cancerous cells may partly explain why glioblastomas are so often hard to beat.
While more study is needed to confirm the role of this process in people with glioblastoma, the findings are an important reminder that treatment advances in oncology may come not only from study of the cancer itself but also by carefully considering the larger context and environments in which tumors grow. The hope is that these intriguing new findings will one day lead to new treatment options for the approximately 13,000 people in the U.S. alone who are diagnosed with glioblastoma each year [2].
References:
[1] GAP43-dependent mitochondria transfer from astrocytes enhances glioblastoma tumorigenicity. Watson DC, Bayik D, Storevik S, Moreino SS, Hjelmeland AB, Hossain JA, Miletic H, Lathia JD et al. Nat Cancer. 2023 May 11. [Published online ahead of print.]
[2] CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. 2018 Oct 1, Neuro Oncol., p. 20(suppl_4):iv1-iv86.
Links:
Glioblastoma (National Center for Advancing Translational Sciences/NIH)
Brain Tumors (National Cancer Institute/NIH)
Justin Lathia Lab (Cleveland Clinic, OH)
Hrvoje Miletic (University of Bergen, Norway)
NIH Support: National Institute of Neurological Disorders and Stroke; National Center for Advancing Translational Sciences; National Cancer Institute; National Institute of Allergy and Infectious Diseases
Posted In: News
Tags: actin filaments, astrocytes, brain cancer, cancer, cancer biology, cell biology, central nervous system cancers, GAPa43, glia, glial cells, glioblastoma, glioblastoma multiforme, mitochondria, mitochondria transfer
Saving Fat for Lean Times
Posted on February 28th, 2023 by Lawrence Tabak, D.D.S., Ph.D.
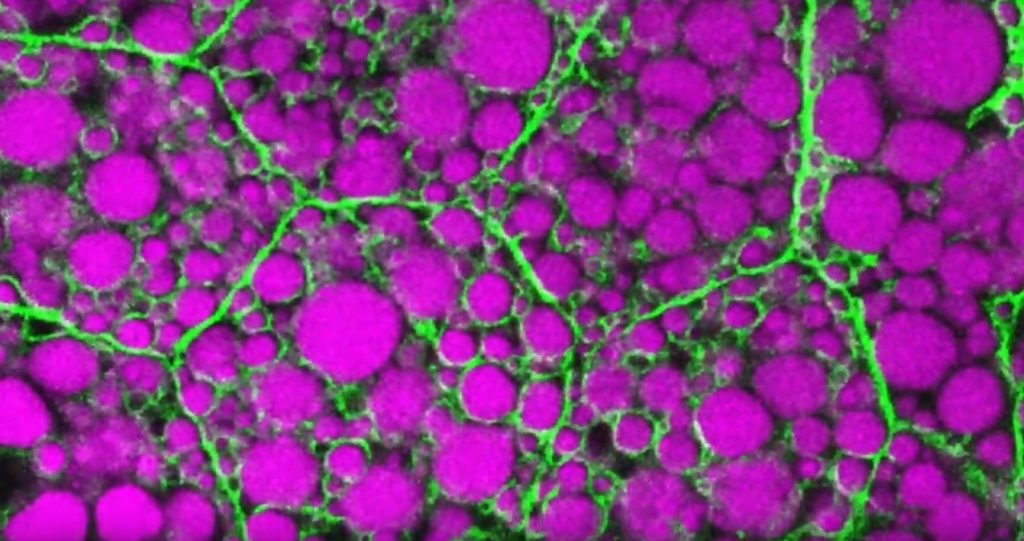
Credit: Rupali Ugrankar, Henne Lab, University of Texas Southwestern Medical Center, Dallas
Humans and all multi-celled organisms, or metazoans, have evolved through millennia into a variety of competing shapes, sizes, and survival strategies. But all metazoans still share lots of intriguing cell biology, including the ability to store excess calories as fat. In fact, many researchers now consider fat-storing cells to be “nutrient sinks,” or good places for the body to stash excess sugars and lipids. Not only can these provide energy needed to survive a future famine, this is a good way to sequester extra molecules that could prove toxic to cells and organs.
Here’s something to think about the next time you skip a meal. Fat-storing cells organize their fat reserves spatially, grouping them into specific pools of lipid types, in order to generate needed energy when food is scarce.
That’s the story behind this striking image taken in a larval fruit fly (Drosophila melanogaster). The image captures fat-storing adipocytes in an organ called a fat body, where a larval fruit fly stores extra nutrients. It’s like the fat tissue in mammals. You can see both large and small lipid droplets (magenta) inside polygon-shaped fat cells, or adipocytes, lined by their plasma membranes (green). But notice that the small lipid droplets are more visibly lined by green, as only these are destined to be saved for later and exported when needed into the fly’s bloodstream.
Working in Mike Henne’s lab at the University of Texas Southwestern Medical Center, Dallas, research associate Rupali Ugrankar discovered how this clever fat-management system works in Drosophila [1]. After either feeding flies high-or-extremely low-calorie diets, Ugrankar used a combination of high-resolution fluorescence confocal microscopy and thin-section transmission electron microscopy to provide a three-dimensional view of adipocytes and their lipid droplets inside.
She observed two distinct sizes of lipid droplets and saw that only the small ones clustered at the cell surface membrane. The adipocytes contorted their membrane inward to grab these small droplets and package them into readily exportable energy bundles.
Ugrankar saw that during times of plenty, a protein machine could fill these small membrane-wrapped fat droplets with lots of triacylglycerol, a high-energy, durable form of fat storage. Their ready access at the surface of the adipocyte allows the fly to balance lipid storage locally with energy release into its blood in times of famine.
Ugrankar’s adeptness at the microscope resulted in this beautiful photo, which was earlier featured in the American Society for Cell Biology’s Green Fluorescent Protein Image and Video Contest. But her work and that of many others help to open a vital window into nutrition science and many critical mechanistic questions about the causes of obesity, insulin resistance, hyperglycemia, and even reduced lifespan.
Such basic research will provide the basis for better therapies to correct these nutrition-related health problems. But the value of basic science must not be forgotten—some of the most important leads could come from a tiny insect in its larval state that shares many aspects of mammalian metabolism.
Reference:
[1] Drosophila Snazarus regulates a lipid droplet population at plasma membrane-droplet contacts in adipocytes. Ugrankar R, Bowerman J, Hariri H, Chandra M, et al. Dev Cell. 2019 Sep 9;50(5):557-572.e5.
Links:
The Interactive Fly (Society for Developmental Biology, Rockville, MD)
Henne Lab (University of Texas Southwestern Medical Center, Dallas)
NIH Support: National Institute of General Medical Sciences
Posted In: Snapshots of Life
Tags: 2019 Green Fluorescent Protein Image and Video Contest, adipocyte, American Society for Cell Biology, basic research, cell biology, Drosophila melanogaster, fat, fat body, fat cells, fat storage, fluorescence microscopy, fruit fly, hyperglycemia, imaging, insulin resistance, lifespan, lipid droplet, lipid storage, lipids, metabolism, model organisms, nutrient sink, obesity, triacylglycerol
Millions of Single-Cell Analyses Yield Most Comprehensive Human Cell Atlas Yet
Posted on May 24th, 2022 by Lawrence Tabak, D.D.S., Ph.D.

There are 37 trillion or so cells in our bodies that work together to give us life. But it may surprise you that we still haven’t put a good number on how many distinct cell types there are within those trillions of cells.
That’s why in 2016, a team of researchers from around the globe launched a historic project called the Human Cell Atlas (HCA) consortium to identify and define the hundreds of presumed distinct cell types in our bodies. Knowing where each cell type resides in the body, and which genes each one turns on or off to create its own unique molecular identity, will revolutionize our studies of human biology and medicine across the board.
Since its launch, the HCA has progressed rapidly. In fact, it has already reached an important milestone with the recent publication in the journal Science of four studies that, together, comprise the first multi-tissue drafts of the human cell atlas. This draft, based on analyses of millions of cells, defines more than 500 different cell types in more than 30 human tissues. A second draft, with even finer definition, is already in the works.
Making the HCA possible are recent technological advances in RNA sequencing. RNA sequencing is a topic that’s been mentioned frequently on this blog in a range of research areas, from neuroscience to skin rashes. Researchers use it to detect and analyze all the messenger RNA (mRNA) molecules in a biological sample, in this case individual human cells from a wide range of tissues, organs, and individuals who voluntarily donated their tissues.
By quantifying these RNA messages, researchers can capture the thousands of genes that any given cell actively expresses at any one time. These precise gene expression profiles can be used to catalogue cells from throughout the body and understand the important similarities and differences among them.
In one of the published studies, funded in part by the NIH, a team co-led by Aviv Regev, a founding co-chair of the consortium at the Broad Institute of MIT and Harvard, Cambridge, MA, established a framework for multi-tissue human cell atlases [1]. (Regev is now on leave from the Broad Institute and MIT and has recently moved to Genentech Research and Early Development, South San Francisco, CA.)
Among its many advances, Regev’s team optimized single-cell RNA sequencing for use on cell nuclei isolated from frozen tissue. This technological advance paved the way for single-cell analyses of the vast numbers of samples that are stored in research collections and freezers all around the world.
Using their new pipeline, Regev and team built an atlas including more than 200,000 single-cell RNA sequence profiles from eight tissue types collected from 16 individuals. These samples were archived earlier by NIH’s Genotype-Tissue Expression (GTEx) project. The team’s data revealed unexpected differences among cell types but surprising similarities, too.
For example, they found that genetic profiles seen in muscle cells were also present in connective tissue cells in the lungs. Using novel machine learning approaches to help make sense of their data, they’ve linked the cells in their atlases with thousands of genetic diseases and traits to identify cell types and genetic profiles that may contribute to a wide range of human conditions.
By cross-referencing 6,000 genes previously implicated in causing specific genetic disorders with their single-cell genetic profiles, they identified new cell types that may play unexpected roles. For instance, they found some non-muscle cells that may play a role in muscular dystrophy, a group of conditions in which muscles progressively weaken. More research will be needed to make sense of these fascinating, but vital, discoveries.
The team also compared genes that are more active in specific cell types to genes with previously identified links to more complex conditions. Again, their data surprised them. They identified new cell types that may play a role in conditions such as heart disease and inflammatory bowel disease.
Two of the other papers, one of which was funded in part by NIH, explored the immune system, especially the similarities and differences among immune cells that reside in specific tissues, such as scavenging macrophages [2,3] This is a critical area of study. Most of our understanding of the immune system comes from immune cells that circulate in the bloodstream, not these resident macrophages and other immune cells.
These immune cell atlases, which are still first drafts, already provide an invaluable resource toward designing new treatments to bolster immune responses, such as vaccines and anti-cancer treatments. They also may have implications for understanding what goes wrong in various autoimmune conditions.
Scientists have been working for more than 150 years to characterize the trillions of cells in our bodies. Thanks to this timely effort and its advances in describing and cataloguing cell types, we now have a much better foundation for understanding these fundamental units of the human body.
But the latest data are just the tip of the iceberg, with vast flows of biological information from throughout the human body surely to be released in the years ahead. And while consortium members continue making history, their hard work to date is freely available to the scientific community to explore critical biological questions with far-reaching implications for human health and disease.
References:
[1] Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Eraslan G, Drokhlyansky E, Anand S, Fiskin E, Subramanian A, Segrè AV, Aguet F, Rozenblatt-Rosen O, Ardlie KG, Regev A, et al. Science. 2022 May 13;376(6594):eabl4290.
[2] Cross-tissue immune cell analysis reveals tissue-specific features in humans. Domínguez Conde C, Xu C, Jarvis LB, Rainbow DB, Farber DL, Saeb-Parsy K, Jones JL,Teichmann SA, et al. Science. 2022 May 13;376(6594):eabl5197.
[3] Mapping the developing human immune system across organs. Suo C, Dann E, Goh I, Jardine L, Marioni JC, Clatworthy MR, Haniffa M, Teichmann SA, et al. Science. 2022 May 12:eabo0510.
Links:
Ribonucleic acid (RNA) (National Human Genome Research Institute/NIH)
Studying Cells (National Institute of General Medical Sciences/NIH)
Regev Lab (Broad Institute of MIT and Harvard, Cambridge, MA)
NIH Support: Common Fund; National Cancer Institute; National Human Genome Research Institute; National Heart, Lung, and Blood Institute; National Institute on Drug Abuse; National Institute of Mental Health; National Institute on Aging; National Institute of Allergy and Infectious Diseases; National Institute of Neurological Disorders and Stroke; National Eye Institute
Tags: autoimmune disorders, cell biology, cell reference maps, cells, connective tissue, genotype-tissue expression, GTEx, HCA, heart disease, Human Cell Atlas, immunology, inflammatory bowel disease, lungs, machine learning, macrophage, messenger RNA, mRNA, muscle, muscular dystrophy, RNA, RNA sequencing, single-cell analysis, single-cell RNA sequencing
NCI Support for Basic Science Paves Way for Kidney Cancer Drug Belzutifan
Posted on January 25th, 2022 by Norman "Ned" Sharpless, M.D., National Cancer Institute

There’s exciting news for people with von Hippel-Lindau (VHL) disease, a rare genetic disorder that can lead to cancerous and non-cancerous tumors in multiple organs, including the brain, spinal cord, kidney, and pancreas. In August 2021, the U.S. Food and Drug Administration (FDA) approved belzutifan (Welireg), a new drug that has been shown in a clinical trial led by National Cancer Institute (NCI) researchers to shrink some tumors associated with VHL disease [1], which is caused by inherited mutations in the VHL tumor suppressor gene.
As exciting as this news is, relatively few people have this rare disease. The greater public health implication of this advancement is for people with sporadic, or non-inherited, clear cell kidney cancer, which is by far the most common subtype of kidney cancer, with more than 70,000 cases and about 14,000 deaths per year. Most cases of sporadic clear cell kidney cancer are caused by spontaneous mutations in the VHL gene.
This advancement is also a great story of how decades of support for basic science through NCI’s scientists in the NIH Intramural Research Program and its grantees through extramural research funding has led to direct patient benefit. And it’s a reminder that we never know where basic science discoveries might lead.
Belzutifan works by disrupting the process by which the loss of VHL in a tumor turns on a series of molecular processes. These processes involve the hypoxia-inducible factor (HIF) transcription factor and one of its subunits, HIF-2α, that lead to tumor formation.
The unraveling of the complex relationship among VHL, the HIF pathway, and cancer progression began in 1984, when Bert Zbar, Laboratory of Immunobiology, NCI-Frederick; and Marston Linehan, NCI’s Urologic Oncology Branch, set out to find the gene responsible for clear cell kidney cancer. At the time, there were no effective treatments for advanced kidney cancer, and 80 percent of patients died within two years.
Zbar and Linehan started by studying patients with sporadic clear cell kidney cancer, but then turned their focus to investigations of people affected with VHL disease, which predisposes a person to developing clear cell kidney cancer. By studying the patients and the genetic patterns of tumors collected from these patients, the researchers hypothesized that they could find genes responsible for kidney cancer.
Linehan established a clinical program at NIH to study and manage VHL patients, which facilitated the genetic studies. It took nearly a decade, but, in 1993, Linehan, Zbar, and Michael Lerman, NCI-Frederick, identified the VHL gene, which is mutated in people with VHL disease. They soon discovered that tumors from patients with sporadic clear cell kidney cancer also have mutations in this gene.
Subsequently, with NCI support, William G. Kaelin Jr., Dana-Farber Cancer Institute, Boston, discovered that VHL is a tumor suppressor gene that, when inactivated, leads to the accumulation of HIF.
Another NCI grantee, Gregg L. Semenza, Johns Hopkins School of Medicine, Baltimore, identified HIF as a transcription factor. And Peter Ratcliffe, University of Oxford, United Kingdom, discovered that HIF plays a role in blood vessel development and tumor growth.
Kaelin and Ratcliffe simultaneously showed that the VHL protein tags a subunit of HIF for destruction when oxygen levels are high. These results collectively answered a very old question in cell biology: How do cells sense the intracellular level of oxygen?
Subsequent studies by Kaelin, with NCI’s Richard Klausner and Linehan, revealed the critical role of HIF in promoting the growth of clear cell kidney cancer. This work ultimately focused on one member of the HIF family, the HIF-2α subunit, as the key mediator of clear cell kidney cancer growth.
The fundamental work of Kaelin, Semenza, and Ratcliffe earned them the 2019 Nobel Prize in Physiology or Medicine. It also paved the way for drug discovery efforts that target numerous points in the pathway leading to clear cell kidney cancer, including directly targeting the transcriptional activity of HIF-2α with belzutifan.
Clinical trials of belzutifan, including several supported by NCI, demonstrated potent anti-cancer activity in VHL-associated kidney cancer, as well as other VHL-associated tumors, leading to the aforementioned recent FDA approval. This is an important development for patients with VHL disease, providing a first-in-class therapy that is effective and well-tolerated.
We believe this is only the beginning for belzutifan’s use in patients with cancer. A number of trials are now studying the effectiveness of belzutifan for sporadic clear cell kidney cancer. A phase 3 trial is ongoing, for example, to look at the effectiveness of belzutifan in treating people with advanced kidney cancer. And promising results from a phase 2 study show that belzutifan, in combination with cabozantinib, a widely used agent to treat kidney cancer, shrinks tumors in patients previously treated for metastatic clear cell kidney cancer [2].
This is a great scientific story. It shows how studies of familial cancer and basic cell biology lead to effective new therapies that can directly benefit patients. I’m proud that NCI’s support for basic science, both intramurally and extramurally, is making possible many of the discoveries leading to more effective treatments for people with cancer.
References:
[1] Belzutifan for Renal Cell Carcinoma in von Hippel-Lindau Disease. Jonasch E, Donskov F, Iliopoulos O, Rathmell WK, Narayan VK, Maughan BL, Oudard S, Else T, Maranchie JK, Welsh SJ, Thamake S, Park EK, Perini RF, Linehan WM, Srinivasan R; MK-6482-004 Investigators. N Engl J Med. 2021 Nov 25;385(22):2036-2046.
[2] Phase 2 study of the oral hypoxia-inducible factor 2α (HIF-2α) inhibitor MK-6482 in combination with cabozantinib in patients with advanced clear cell renal cell carcinoma (ccRCC). Choueiri TK et al. J Clin Oncol. 2021 Feb 20;39(6_suppl): 272-272.
Links:
Von Hippel-Lindau Disease (Genetic and Rare Diseases Information Center/National Center for Advancing Translational Sciences/NIH)
The Long Road to Understanding Kidney Cancer (Intramural Research Program/NIH)
[Note: Acting NIH Director Lawrence Tabak has asked the heads of NIH’s institutes and centers to contribute occasional guest posts to the blog as a way to highlight some of the cool science that they support and conduct. This is the first in the series of NIH institute and center guest posts that will run until a new permanent NIH director is in place.]
Posted In: Generic
Tags: 2019 Nobel Prize in Physiology or Medicine, basic research, belzutifan, cancer, cancer drugs, cell biology, clear cell kidney cancer, clinical trials, FDA, genetics, HIF, kidney cancer, NCI, NCI-MATCH trial, oncology, rare disease, tumor suppressor gene, VHL, von Hippel-Lindau disease, Welireg
New Microscope Technique Provides Real-Time 3D Views
Posted on September 30th, 2021 by Dr. Francis Collins
Most of the “cool” videos shared on my blog are borne of countless hours behind a microscope. Researchers must move a biological sample through a microscope’s focus, slowly acquiring hundreds of high-res 2D snapshots, one painstaking snap at a time. Afterwards, sophisticated computer software takes this ordered “stack” of images, calculates how the object would look from different perspectives, and later displays them as 3D views of life that can be streamed as short videos.
But this video is different. It was created by what’s called a multi-angle projection imaging system. This new optical device requires just a few camera snapshots and two mirrors to image a biological sample from multiple angles at once. Because the device eliminates the time-consuming process of acquiring individual image slices, it’s up to 100 times faster than current technologies and doesn’t require computer software to construct the movie. The kicker is that the video can be displayed in real time, which isn’t possible with existing image-stacking methods.
The video here shows two human melanoma cells, rotating several times between overhead and side views. You can see large amounts of the protein PI3K (brighter orange hues indicate higher concentrations), which helps some cancer cells divide and move around. Near the cell’s perimeter are small, dynamic surface protrusions. PI3K in these “blebs” is thought to help tumor cells navigate and survive in foreign tissues as the tumor spreads to other organs, a process known as metastasis.
The new multi-angle projection imaging system optical device was described in a paper published recently in the journal Nature Methods [1]. It was created by Reto Fiolka and Kevin Dean at the University of Texas Southwestern Medical Center, Dallas.
Like most technology, this device is complicated. Rather than the microscope and camera doing all the work, as is customary, two mirrors within the microscope play a starring role. During a camera exposure, these mirrors rotate ever so slightly and warp the acquired image in such a way that successive, unique perspectives of the sample magically come into view. By changing the amount of warp, the sample appears to rotate in real-time. As such, each view shown in the video requires only one camera snapshot, instead of acquiring hundreds of slices in a conventional scheme.
The concept traces to computer science and an algorithm called the shear warp transform method. It’s used to observe 3D objects from different perspectives on a 2D computer monitor. Fiolka, Dean, and team found they could implement a similar algorithm optically for use with a microscope. What’s more, their multi-angle projection imaging system is easy-to-use, inexpensive, and can be converted for use on any camera-based microscope.
The researchers have used the device to view samples spanning a range of sizes: from mitochondria and other tiny organelles inside cells to the beating heart of a young zebrafish. And, as the video shows, it has been applied to study cancer and other human diseases.
In a neat, but also scientifically valuable twist, the new optical method can generate a virtual reality view of a sample. Any microscope user wearing the appropriately colored 3D glasses immediately sees the objects.
While virtual reality viewing of cellular life might sound like a gimmick, Fiolka and Dean believe that it will help researchers use their current microscopes to see any sample in 3D—offering the chance to find rare and potentially important biological events much faster than is possible with even the most advanced microscopes today.
Fiolka, Dean, and team are still just getting started. Because the method analyzes tissue very quickly within a single image frame, they say it will enable scientists to observe the fastest events in biology, such as the movement of calcium throughout a neuron—or even a whole bundle of neurons at once. For neuroscientists trying to understand the brain, that’s a movie they will really want to see.
Reference:
[1] Real-time multi-angle projection imaging of biological dynamics. Chang BJ, Manton JD, Sapoznik E, Pohlkamp T, Terrones TS, Welf ES, Murali VS, Roudot P, Hake K, Whitehead L, York AG, Dean KM, Fiolka R. Nat Methods. 2021 Jul;18(7):829-834.
Links:
Metastatic Cancer: When Cancer Spreads (National Cancer Institute)
Fiolka Lab (University of Texas Southwestern Medical Center, Dallas)
Dean Lab (University of Texas Southwestern)
Microscopy Innovation Lab (University of Texas Southwestern)
NIH Support: National Cancer Institute; National Institute of General Medical Sciences
Posted In: Cool Videos
Tags: 3D imaging, basic research, brain, cancer, cancer biology, cancer metastasis, cell biology, cells, imaging, melanoma, microscopy, multi-angle projection imaging system optical device, neurons, PI3K, shear warp transform method
The Hidden Beauty of Intestinal Villi
Posted on July 22nd, 2021 by Dr. Francis Collins

Credit: Amy Engevik, Medical University of South Carolina, Charleston.
The human small intestine, though modest in diameter and folded compactly to fit into the abdomen, is anything but small. It measures on average about 20 feet from end to end and plays a big role in the gastrointestinal tract, breaking down food and drink from the stomach to absorb the water and nutrients.
Also anything but small is the total surface area of the organ’s inner lining, where millions of U-shaped folds in the mucosal tissue triple the available space to absorb the water and nutrients that keep our bodies nourished. If these folds, packed with finger-like absorptive cells called villi, were flattened out, they would be the size of a tennis court!
That’s what makes this this microscopic image so interesting. It shows in cross section the symmetrical pattern of the villi (its cells outlined by yellow) that pack these folds. Each cell’s nucleus contains DNA (teal), and the villi themselves are fringed by thousands of tiny bristles, called microvilli (magenta), which are too small to see individually here. Collectively, microvilli make up an absorptive surface, called the brush border, where digested nutrients in the fluid passing through the intestine can enter cells via transport channels.
Amy Engevik, a researcher at the Medical University of South Carolina, Charleston, took this snapshot to show what a healthy intestinal cellular landscape looks like in a young mouse. The Engevik lab studies the dynamic movement of ions, water, and proteins in the intestine—a process that goes wrong in humans born with a rare disorder called microvillus inclusion disease (MVID).
MVID causes chronic gastrointestinal problems in newborn babies, due to defects in a protein that transports various cellular components. Because they cannot properly absorb nutrition from food, these tiny patients require intravenous feeding almost immediately, which carries a high risk for sepsis and intestinal injury.
Engevik and her team study this disease using a mouse model that replicates many of the characteristics of the disorder in humans [1]. Interestingly, when Engevik gets together with her family, she isn’t the only one talking about MVID and villi. Her two sisters, Mindy and Kristen, also study the basic science of gastrointestinal disorders! Instead of sibling rivalry, though, this close alliance has strengthened the quality of her research, says Amy, who is the middle child.
Beyond advancing science and nurturing sisterhood in science, Engevik’s work also captured the fancy of the judges for the Federation of American Societies for Experimental Biology’s annual BioArt Scientific Image and Video Competition. Her image was one of 10 winners announced in December 2020.
Because multiple models are useful for understanding fundamentals of diseases like MVID, Engevik has also developed a large-animal model (pig) that has many features of the human disease [2]. She hopes that her efforts will help to improve our understanding of MVID and other digestive diseases, as well as lead to new, potentially life-saving treatments for babies suffering from MVID.
References:
[1] Loss of MYO5B Leads to reductions in Na+ absorption with maintenance of CFTR-dependent Cl- secretion in enterocytes. Engevik AC, Kaji I, Engevik MA, Meyer AR, Weis VG, Goldstein A, Hess MW, Müller T, Koepsell H, Dudeja PK, Tyska M, Huber LA, Shub MD, Ameen N, Goldenring JR. Gastroenterology. 2018 Dec;155(6):1883-1897.e10.
[2] Editing myosin VB gene to create porcine model of microvillus inclusion disease, with microvillus-lined inclusions and alterations in sodium transporters. Engevik AC, Coutts AW, Kaji I, Rodriguez P, Ongaratto F, Saqui-Salces M, Medida RL, Meyer AR, Kolobova E, Engevik MA, Williams JA, Shub MD, Carlson DF, Melkamu T, Goldenring JR. Gastroenterology. 2020 Jun;158(8):2236-2249.e9.
Links:
Microvillus inclusion disease (Genetic and Rare Diseases Center/NIH)
Digestive Diseases (National Institute of Diabetes and Digestive and Kidney Diseases/NIH)
Amy Engevik (Medical University of South Carolina, Charleston)
Podcast: A Tale of Three Sisters featuring Drs. Mindy, Amy, and Kristen Engevik (The Immunology Podcast, April 29, 2021)
BioArt Scientific Image and Video Competition (Federation of American Societies for Experimental Biology, Bethesda, MD)
NIH Support: National Institute of Diabetes and Digestive and Kidney Diseases
Posted In: Snapshots of Life
Tags: animal models, brush border, cell biology, digestion, digestive diseases, digestive tract, FASEB Bioart 2020, gastroenterology, gastrointestinal tract, immunology, intestine, microvilli, microvillus inclusion disease, mouse, MVID, pig, rare diseaes, small intestine, villi
Single-Cell Study Offers New Clue into Causes of Cystic Fibrosis
Posted on May 27th, 2021 by Dr. Francis Collins

Credit: Carraro G, Nature, 2021
More than 30 years ago, I co-led the Michigan-Toronto team that discovered that cystic fibrosis (CF) is caused by an inherited misspelling in the cystic fibrosis transmembrane conductance regulator (CFTR) gene [1]. The CFTR protein’s normal function on the surface of epithelial cells is to serve as a gated channel for chloride ions to pass in and out of the cell. But this function is lost in individuals for whom both copies of CFTR are misspelled. As a consequence, water and salt get out of balance, leading to the production of the thick mucus that leaves people with CF prone to life-threatening lung infections.
It took three decades, but that CFTR gene discovery has now led to the development of a precise triple drug therapy that activates the dysfunctional CFTR protein and provides major benefit to most children and adults with CF. But about 10 percent of individuals with CF have mutations that result in the production of virtually no CFTR protein, which means there is nothing for current triple therapy to correct or activate.
That’s why more basic research is needed to tease out other factors that contribute to CF and, if treatable, could help even more people control the condition and live longer lives with less chronic illness. A recent NIH-supported study, published in the journal Nature Medicine [2], offers an interesting basic clue, and it’s visible in the image above.
The healthy lung tissue (left) shows a well-defined and orderly layer of ciliated cells (green), which use hair-like extensions to clear away mucus and debris. Running closely alongside it is a layer of basal cells (outlined in red), which includes stem cells that are essential for repairing and regenerating upper airway tissue. (DNA indicating the position of cell is stained in blue).
In the CF-affected airways (right), those same cell types are present. However, compared to the healthy lung tissue, they appear to be in a state of disarray. Upon closer inspection, there’s something else that’s unusual if you look carefully: large numbers of a third, transitional cell subtype (outlined in red with green in the nucleus) that combines properties of both basal stem cells and ciliated cells, which is suggestive of cells in transition. The image below more clearly shows these cells (yellow arrows).

Credit: Carraro G, Nature, 2021
The increased number of cells with transitional characteristics suggests an unsuccessful attempt by the lungs to produce more cells capable of clearing the mucus buildup that occurs in airways of people with CF. The data offer an important foundation and reference for continued study.
These findings come from a team led by Kathrin Plath and Brigitte Gomperts, University of California, Los Angeles; John Mahoney, Cystic Fibrosis Foundation, Lexington, MA; and Barry Stripp, Cedars-Sinai, Los Angeles. Together with their lab members, they’re part of a larger research team assembled through the Cystic Fibrosis Foundation’s Epithelial Stem Cell Consortium, which seeks to learn how the disease changes the lung’s cellular makeup and use that new knowledge to make treatment advances.
In this study, researchers analyzed the lungs of 19 people with CF and another 19 individuals with no evidence of lung disease. Those with CF had donated their lungs for research in the process of receiving a lung transplant. Those with healthy lungs were organ donors who died of other causes.
The researchers analyzed, one by one, many thousands of cells from the airway and classified them into subtypes based on their distinctive RNA patterns. Those patterns indicate which genes are switched on or off in each cell, as well as the degree to which they are activated. Using a sophisticated computer-based approach to sift through and compare data, the team created a comprehensive catalog of cell types and subtypes present in healthy airways and in those affected by CF.
The new catalogs also revealed that the airways of people with CF had alterations in the types and proportions of basal cells. Those differences included a relative overabundance of cells that appeared to be transitioning from basal stem cells into the specialized ciliated cells, which are so essential for clearing mucus from the lungs.
We are not yet at our journey’s end when it comes to realizing the full dream of defeating CF. For the 10 percent of CF patients who don’t benefit from the triple-drug therapy, the continuing work to find other treatment strategies should be encouraging news. Keep daring to dream of breathing free. Through continued research, we can make the story of CF into history!
References:
[1] Identification of the cystic fibrosis gene: chromosome walking and jumping. Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. Science.1989 Sep 8;245(4922):1059-65.
[2] Transcriptional analysis of cystic fibrosis airways at single-cell resolution reveals altered epithelial cell states and composition. Carraro G, Langerman J, Sabri S, Lorenzana Z, Purkayastha A, Zhang G, Konda B, Aros CJ, Calvert BA, Szymaniak A, Wilson E, Mulligan M, Bhatt P, Lu J, Vijayaraj P, Yao C, Shia DW, Lund AJ, Israely E, Rickabaugh TM, Ernst J, Mense M, Randell SH, Vladar EK, Ryan AL, Plath K, Mahoney JE, Stripp BR, Gomperts BN. Nat Med. 2021 May;27(5):806-814.
Links:
Cystic Fibrosis (National Heart, Lung, and Blood Institute/NIH)
Kathrin Plath (University of California, Los Angeles)
Brigitte Gomperts (UCLA)
Stripp Lab (Cedars-Sinai, Los Angeles)
Cystic Fibrosis Foundation (Lexington, MA)
Epithelial Stem Cell Consortium (Cystic Fibrosis Foundation, Lexington, MA)
NIH Support: National Heart, Lung, and Blood Institute; National Institute of Diabetes and Digestive and Kidney Diseases; National Institute of General Medical Sciences; National Cancer Institute; National Center for Advancing Translational Sciences
Posted In: News
Tags: basal cells, cell biology, CF, CFTR, ciliated cells, cystic fibrosis, Cystic Fibrosis Foundation, epithelial cells, Epithelial Stem Cell Consortium, gene expression, lungs, mucus, rare disease, RNA, single cell analysis, single cell sequencing, transitional cell subtype, upper airway
Capturing Viral Shedding in Action
Posted on April 28th, 2020 by Dr. Francis Collins

Credit: Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, Hamilton, MT
You’ve probably seen some amazing high-resolution images of SARS-CoV-2, the novel coronavirus that causes COVID-19, on television and the web. What you might not know is that many of these images, including the ones shown here, were produced at Rocky Mountain Laboratories (RML), a part of NIH’s National Institute of Allergy and Infectious Diseases (NIAID) that’s located in the small Montana town of Hamilton.
The head of RML’s Electron Microscopy Unit, Elizabeth Fischer, was the researcher who took this portrait of SARS-CoV-2. For more than 25 years, Fischer has snapped stunning images of dangerous viruses and microbes, including some remarkable shots of the deadly Ebola virus. She also took some of the first pictures of the coronavirus that causes Middle East respiratory syndrome (MERS), which arose from camels and continues to circulate at low levels in people.
The NIAID facility uses a variety of microscopy techniques, including state-of-the-art cryo-electron microscopy (cryo-EM). But the eye-catching image you see here was taken with a classic scanning electron microscope (SEM).
SEM enables visualization of particles, including viruses, that are too small to be seen with traditional light microscopy. It does so by focusing electrons, instead of light, into a beam that scans the surface of a sample that’s first been dehydrated, chemically preserved, and then coated with a thin layer of metal. As electrons bounce off the sample’s surface, microscopists such as Fischer are able to capture its precise topology. The result is a gray-scale micrograph like the one you see above on the left. To make the image easier to interpret, Fischer hands the originals off to RML’s Visual Medical Arts Department, which uses colorization to make key features pop like they do in the image on the right.
So, what exactly are you seeing in this image? The orange-brown folds and protrusions are part of the surface of a single cell that’s been infected with SARS-CoV-2. This particular cell comes from a commonly studied primate kidney epithelial cell line. The small, blue spheres emerging from the cell surface are SARS-CoV-2 particles.
This picture is quite literally a snapshot of viral shedding, a process in which viral particles are released from a dying cell. This image gives us a window into how devastatingly effective SARS-CoV-2 appears to be at co-opting a host’s cellular machinery: just one infected cell is capable of releasing thousands of new virus particles that can, in turn, be transmitted to others.
While capturing a fixed sample on the microscope is fairly straightforward for a pro like Fischer, developing a sample like this one involves plenty of behind-the-scenes trial and error by NIAID investigators. As you might imagine, to see the moment that viruses emerge from an infected cell, you have to get the timing just right.
By capturing many shots of the coronavirus using the arsenal of microscopes available at RML and elsewhere, researchers are learning more every day about how SARS-CoV-2 enters a cell, moves inside it, and then emerges to infect other cells. In addition to advancing scientific knowledge, Fischer notes that images like these also hold the remarkable power to make an invisible enemy visible to the world at large.
Making SARS-CoV-2 tangible helps to demystify the challenges that all of us now face as a result of the COVID-19 pandemic. The hope is it will encourage each and every one of us to do our part to fight it, whether that means digging into the research, working on the front lines, or staying at home to prevent transmission and flatten the curve. And, if you could use some additional inspiration, don’t miss the NIAID’s image gallery on Flickr, which includes some of Fischer’s finest work.
Links:
Coronavirus (COVID-19) (NIH)
Rocky Mountain Laboratories (National Institute of Allergy and Infectious Diseases/NIH)
Elizabeth Fischer (National Institute of Allergy and Infectious Diseases/NIH)
NIH Support: National Institute of Allergy and Infectious Diseases
Posted In: Snapshots of Life
Tags: cell biology, coronavirus disease 2019, COVID-19, Ebola, Ebola virus, imaging, infectious diseases, MERS, microscopy, Middle East respiratory syndrome, novel coronavirus, pandemic, Rocky Mountain Laboratories, Rocky Mountain Labs, SARS-CoV-2, scanning electron microscopy, SEM, Vero cells, viral shedding, virus