diagnostics – NIH Director's Blog (original) (raw)
The Chemistry Clicked: Two NIH-Supported Researchers Win 2022 Nobel Prize in Chemistry
Posted on October 11th, 2022 by Lawrence Tabak, D.D.S., Ph.D.

Through the years, NIH has supported a total of 169 researchers who have received or shared 101 Nobel Prizes. That’s quite a testament to the world-leading science that NIH pursues and its continued impact on improving human health and well-being.
Those numbers include the news late last week that the 2022 Nobel Prize in Chemistry was shared by two long-time grantees for their work on a transformative scientific approach known as “click chemistry.” This form of chemistry has made it possible for researchers to snap together, like LEGO pieces, molecular building blocks to form hybrid biomolecules, often with easy-to-track imaging agents attached. Not only has click chemistry expanded our ability to explore the molecular underpinnings of a wide range of biological processes, but it has provided us with new tools for developing drugs, diagnostics, and a wide array of “smart” materials.
For K. Barry Sharpless, Scripps Research, La Jolla, CA, October 5, 2022 marked the second time that he’s received an early-morning congratulatory call from The Royal Swedish Academy of Sciences. The first such call came in 2001, when Sharpless got the news that he was a co-winner of the Nobel Prize in Chemistry for his discovery of asymmetric catalytic reactions.
This time around, Sharpless was recognized for his groundbreaking studies in the mid-1990s with click chemistry, a term that he coined himself. His initial work established click chemistry as a fast-and-reliable way to attach molecules of interest in the lab [1]. He and co-recipient Morten Meldal, University of Copenhagen, Denmark, who is not funded by NIH, then independently introduced a copper-catalyzed click that further refined the chemistry and helped popularize it across biology and the material sciences [2,3].
For Carolyn R. Bertozzi of Stanford University, Palo Alto, CA, it is her first Nobel. Bertozzi was recognized for expanding the use of click chemistry with so-called bioorthogonal chemistry, which is a copper-free version of the approach that can be used inside living cells without the risk of metal-associated toxicities [4,5].
Bertozzi’s work has been especially interesting to me because of her focus on glycans, which I’ve studied throughout my career. Glycans are the carbohydrate molecules that coat the surfaces of our cells and most secreted proteins. They are essential to life, and, in higher organisms, play fundamental roles in basic processes such as metabolism, immunity, and cellular communication.
Glycans also remain poorly understood, largely because, until recently, they have been so difficult for basic scientists to study with traditional techniques. That has changed with development of new tools to study glycans and the enzymes that assemble them. My long-time collaborator, Kelly Ten Hagen, a senior investigator at NIH’s National Institute of Dental and Craniofacial Research, and I collaborated with Carolyn on identifying small molecules that inhibit the enzyme responsible for the first step in mucin-type O-glycosylation [6]
In the early 2000s, Bertozzi and her team introduced bioorthogonal chemistry, which enabled researchers to label glycans and visualize them in a range of cells and living organisms. Her team’s pioneering approach quickly became an essential tool in basic science labs around the world that study glycans, leading to a number of stunning discoveries that would have otherwise been difficult or impossible.
For clinical researchers, click chemistry has emerged as a workhorse in drug discovery and the improved targeting of cancer chemotherapies and other small-molecule drugs. The approach also is being used to improve delivery of antibody-based therapies and to create new biomaterials. Meanwhile, in the material sciences, click chemistry has been used to solve a number of problems in working with polymers and to expand their industrial uses.
Click chemistry is an excellent example of how advances in basic science can build the foundation for a wide range of practical applications, including those aimed at improving human health. It also highlights the value of strong, sustained public funding for fundamental research, and NIH is proud to have supported Sharpless continuously since 1975 and Bertozzi since 1999. I send my sincere congratulations to both of these most-deserving scientists.
References:
[1] Click Chemistry: Diverse chemical function from a few good reactions. Kolb, HC, Finn, MG, Sharpless, KB. Angew. Chem. Int. Ed. 2001, 40 (11), 2004–2021
[2] A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “Llgation” of azides and terminal alkynes. Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. Int. Ed. 2002, 41 (14), 2596–2599.
[3] Peptidotriazoles on solid phase: [1,2,3]-Triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. Tornøe CW, Sengeløv H, Meldal M. J. Org. Chem. 2002, 67 (9), 3057–3064.
[4] A strain-promoted [3 + 2] azide−alkyne cycloaddition for covalent modification of biomolecules in living systems. Agard NJ, Prescher JA, Bertozzi CR. J. Am. Chem. Soc. 2004, 126 (46), 15046–15047
[5] In vivo imaging of membrane associated glycans in developing zebrafish. Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science 2008, 320 (5876), 664–667.
[6] Small molecule inhibitors of mucin-type O-glycosylation from a uridine-based library. Hang, HC, Yu, C, Ten Hagen, KG, Tian, E, Winans, KA, Tabak, LA, Bertozzi, Chem Biol. 2004 Jul;11(7):1009-1016.
Links:
The Nobel Prize in Chemistry 2022 (The Royal Swedish Academy of Sciences, Stockholm)
Video: Announcement of the 2022 Nobel Prize in Chemistry (YouTube)
Click Chemistry and Bioorthogonal Chemistry (The Royal Swedish Academy of Sciences)
Sharpless Lab (Scripps Research, La Jolla, CA)
Bertozzi Group (Stanford University, Palo Alto, CA)
NIH Support:
K. Barry Sharpless: National Institute of General Medical Sciences
Carolyn R. Bertozzi: National Cancer Institute; National Institute of Allergy and Infectious Diseases; National Institute of General Medical Sciences
Posted In: News
Tags: 2022 Nobel Prize in Chemistry, basic research, basic science, bioorthogonal chemistry, Carolyn Bertozzi, chemistry, click chemistry, copper-catalyzed click chemistry, diagnostics, drug development, glycans, imaging, K. Barry Sharpless, materials science, Morton Meldal, Nobel Laureate, Nobel Prize, O-glycosylation, smart materials, The Royal Swedish Academy, zebrafish
RADx Initiative: Bioengineering for COVID-19 at Unprecedented Speed and Scale
Posted on May 3rd, 2022 by Bruce J. Tromberg, Ph.D., National Institute of Biomedical Imaging and Bioengineering

Credit: Africa Studio/Shutterstock; Quidel Corporation, San Diego, CA
As COVID-19 rapidly expanded throughout the world in April 2020, many in the biomedical technology community voiced significant concerns about the lack of available diagnostic tests. At that time, testing for SARS-CoV-2, the coronavirus that causes COVID-19, was conducted exclusively in clinical laboratories by order of a health-care provider. “Over the counter” (OTC) tests did not exist, and low complexity point of care (POC) platforms were rare. Fewer than 8 million tests were performed in the U.S. that month, and it was clear that we needed a radical transformation to make tests faster and more accessible.
By February 2022, driven by the Omicron variant surge, U.S. capacity had increased to a new record of more than 1.2 billion tests in a single month. Remarkably, the overwhelming majority of these—more than 85 percent—were “rapid tests” conducted in home and POC settings.
The story behind this practice-changing, “test-at-home” transformation is deeply rooted in technologic and manufacturing innovation. The NIH’s National Institute of Biomedical Imaging and Bioengineering (NIBIB), working collaboratively with multiple partners across NIH, government, academia, and the private sector, has been privileged to play a leading role in this effort via the Rapid Acceleration of Diagnostics (RADx®) initiative. On this two-year anniversary of RADx, we take a brief look back at its formation, impact, and potential for future growth.
On April 24, 2020, Congress recognized that testing was an urgent national need and appropriated $1.5 billion to NIH via an emergency supplement [1]. The goal was to substantially increase the number, type, and availability of diagnostic tests in only five to six months. Since the “normal” commercialization cycle for this type of diagnostic technology is typically more than five years, we needed an entirely new approach . . . fast.
The RADx initiative was launched just five days after that challenging Congressional directive [2]. Four NIH RADx programs were eventually created to support technology development and delivery, with the goal of matching test performance with community needs [3].The first two programs, RADx Tech and RADx Advanced Technology Platforms (ATP), were developed by NIBIB and focused on innovation for rapidly creating, scaling up, and deploying new technologies.
RADx Tech is built around NIBIB’s Point of Care Technologies Research Network (POCTRN) and includes core activities for technology review, test validation, clinical studies, regulatory authorization, and test deployment. Overall, the RADx Tech network includes approximately 900 participants from government, academia, and the private sector with unique capabilities and resources designed to decrease inherent risk and guide technologies from design and development to fully disseminated commercial products.
At the core of RADx Tech operations is the “innovation funnel” rapid review process, popularized as a shark tank [4]. A total of 824 complete applications were submitted during two open calls in a four-month period, beginning April 2020 and during a one-month period in June 2021. Forty-seven projects received phase 1 funding to validate and lower the inherent risk of developing these technologies. Meanwhile, 50 companies received phase 2 contracts to support FDA authorization studies and manufacturing expansion [5]
Beyond test development, RADx Tech has evolved to become a key contributor to the U.S. COVID-19 response. The RADx Independent Test Assessment Program (ITAP) was launched in October 2021 to accelerate regulatory authorization of new tests as a joint effort with the Food and Drug Administration (FDA) [6]. The ITAP acquires analytical and clinical performance data and works closely with FDA and manufacturers to shave weeks to months off the time it normally takes to receive Emergency Use Authorization (EUA).
The RADx Tech program also created a Variant Task Force to monitor the performance of tests against each new coronavirus “variant of concern” that emerges. This helps to ensure that marketed tests continue to remain effective. Other innovative RADx Tech projects include Say Yes! Covid Test, the first online free OTC test distribution program, and Project Rosa, which conducts real-time variant tracking across the country [7].
RADx Tech, by any measure, has exceeded even the most-optimistic expectations. In two years, RADx Tech-supported companies have received 44 EUAs and added approximately 2 billion tests and test products to the U.S. capacity. These remarkable numbers have steadily increased from more than16 million tests in September 2020, just five months after the program was established [8].
RADx Tech has also made significant contributions to the distribution of 1 billion free OTC tests via the government site, COVID.gov/tests. It has also provided critical guidance on serial testing and variants that have improved test performance and changed regulatory practice [9,10]. In addition, the RADx Mobile Application Reporting System (RADx MARS) reduces barriers to test reporting and test-to-treat strategies’ The latter offers immediate treatment options via telehealth or a POC location whenever a positive test result is reported. Finally, the When to Test website provides critical guidance on when and how to test for individuals, groups, and communities.
As we look to the future, RADx Tech has enormous potential to impact the U.S. response to other pathogens, diseases, and future pandemics. Major challenges going forward include improving home tests to work as well as lab platforms and building digital health networks for capturing and reporting test results to public health officials [11].
A recent editorial published in the journal Nature Biotechnology noted, “RADx has spawned a phalanx of diagnostic products to market in just 12 months. Its long-term impact on point of care, at-home, and population testing may be even more profound [12].” We are now poised to advance a new wave of precision medicine that’s led by innovative diagnostic technologies. It represents a unique opportunity to emerge stronger from the pandemic and achieve long-term impact.
References:
[1] Public Law 116 -139—Paycheck Protection Program and Health Care Enhancement Act.
[2] NIH mobilizes national innovation initiative for COVID-19 diagnostics, NIH news release, April 29, 2020.
[3] Rapid scaling up of Covid-19 diagnostic testing in the United States—The NIH RADx Initiative. Tromberg BJ, Schwetz TA, Pérez-Stable EJ, Hodes RJ, Woychik RP, Bright RA, Fleurence RL, Collins FS. N Engl J Med. 2020 Sep 10;383(11):1071-1077.
[4] We need more covid-19 tests. We propose a ‘shark tank’ to get us there. Alexander L. and Blunt R., Washington Post, April 20, 2020.
[5] RADx® Tech/ATP dashboard, National Institute of Biomedical Imaging and Bioengineering, NIH.
[6] New HHS actions add to Biden Administration efforts to increase access to easy-to-use over-the-counter COVID-19 tests. U.S. Department of Health and Human Services Press Office, October 25, 2021.
[7] A method for variant agnostic detection of SARS-CoV-2, rapid monitoring of circulating variants, detection of mutations of biological significance, and early detection of emergent variants such as Omicron. Lai E, et al. medRxiV preprint, January 9, 2022.
[9] Longitudinal assessment of diagnostic test performance over the course of acute SARS-CoV-2 infection. Smith RL, et al. J Infect Dis. 2021 Sep 17;224(6):976-982.
[10] Comparison of rapid antigen tests’ performance between Delta (B.1.61.7; AY.X) and Omicron (B.1.1.529; BA1) variants of SARS-CoV-2: Secondary analysis from a serial home self-testing study. Soni A, et al. MedRxiv preprint, March 2, 2022.
[11] Reporting COVID-19 self-test results: The next frontier. Health Affairs, Juluru K., et al. Health Affairs, February 11, 2022.
[12] Radical solutions. Nat Biotechnol. 2021 Apr;39(4):391.
Links:
Get Free At-Home COVID Tests (COVID.gov)
When to Test (Consortia for Improving Medicine with Innovation & Technology, Boston)
RADx Programs (NIH)
RADx® Tech and ATP Programs (National Institute of Biomedical Imaging and Biomedical Engineering/NIH)
Independent Test Assessment Program (NIBIB)
Mobile Application Reporting through Standards (NIBIB)
Point-of-Care Technologies Research Network (POCTRN) (NIBIB)
[Note: Acting NIH Director Lawrence Tabak has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the eighth in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.]
Posted In: Generic
Tags: bioengineering, Congress, COVID-19, COVID-19 testing, COVID.gov/tests, diagnostic tests, diagnostics, EUA, FDA, home diagnostics, home tests, innovation funnel, NIBIB, novel coronavirus, Omicron variant, OTC tests, pandemic, POC tests, POCTRN, point of care, precision medicine, public-private partnership, RADx, RADx MARS, RADx-ATP, RADx-tech, Rapid Acceleration of Diagnostics Initiative, SARS-CoV-2, Say Yes! COVID Test, Shark Tank, telehealth, tests, When to Test
A More Precise Way to Knock Out Skin Rashes
Posted on April 26th, 2022 by Lawrence Tabak, D.D.S., Ph.D.
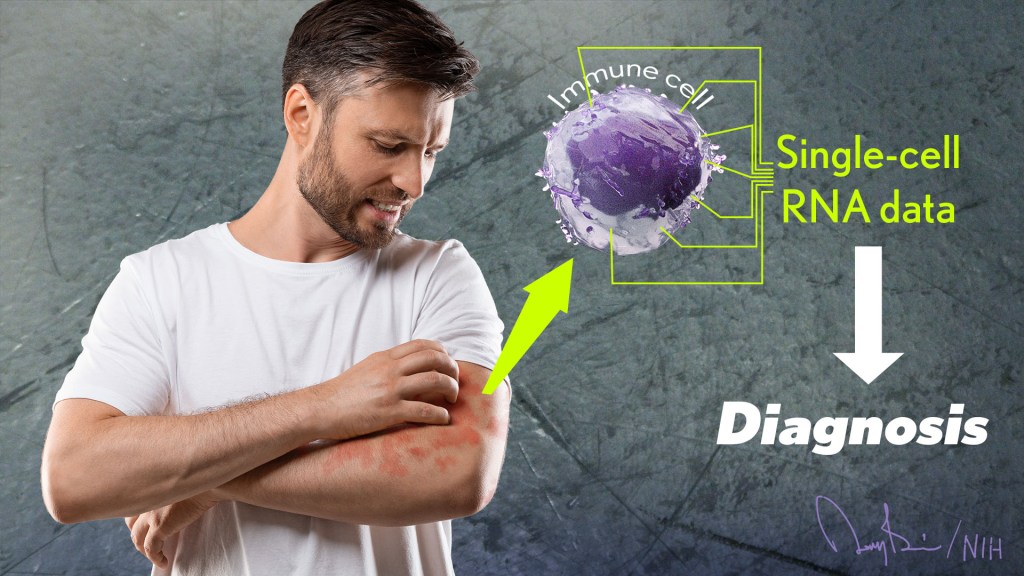
The NIH is committed to building a new era in medicine in which the delivery of health care is tailored specifically to the individual person, not the hypothetical average patient as is now often the case. This new era of “precision medicine” will transform care for many life-threatening diseases, including cancer and chronic kidney disease. But what about non-life-threatening conditions, like the aggravating rash on your skin that just won’t go away?
Recently, researchers published a proof-of-principle paper in the journal Science Immunology demonstrating just how precision medicine for inflammatory skin rashes might work [1]. While more research is needed to build out and further refine the approach, the researchers show it’s now technologically possible to extract immune cells from a patient’s rash, read each cell’s exact inflammatory features, and relatively quickly match them online to the right anti-inflammatory treatment to stop the rash.
The work comes from a NIH-funded team led by Jeffrey Cheng and Raymond Cho, University of California, San Francisco. The researchers focused their attention on two inflammatory skin conditions: atopic dermatitis, the most common type of eczema, which flares up periodically to make skin red and itchy, and psoriasis vulgaris. Psoriasis causes skin cells to build up and form a scaly rash and dry, itchy patches. Together, atopic dermatitis and psoriasis vulgaris affect about 10 percent of U.S. adults.
While the rashes caused by the two conditions can sometimes look similar, they are driven by different sets of immune cells and underlying inflammatory responses. For that reason, distinct biologic therapies, based on antibodies and proteins made from living cells, are now available to target and modify the specific immune pathways underlying each condition.
While biologic therapies represent a major treatment advance for these and other inflammatory conditions, they can miss their targets. Indeed, up to half of patients don’t improve substantially on biologics. Part of the reason for that lack of improvement is because doctors don’t have the tools they need to make firm diagnoses based on what precisely is going on in the skin at the molecular and cellular levels.
To learn more in the new study, the researchers isolated immune cells, focusing primarily on T cells, from the skin of 31 volunteers. They then sequenced the RNA of each cell to provide a telltale portrait of its genomic features. This “single-cell analysis” allowed them to capture high-resolution portraits of 41 different immune cell types found in individual skin samples. That’s important because it offers a much more detailed understanding of changes in the behavior of various immune cells that might have been missed in studies focused on larger groupings of skin cells, representing mixtures of various cell types.
Of the 31 volunteers, seven had atopic dermatitis and eight had psoriasis vulgaris. Three others were diagnosed with other skin conditions, while six had an indeterminate rash with features of both atopic dermatitis and psoriasis vulgaris. Seven others were healthy controls.
The team produced molecular signatures of the immune cells. The researchers then compared the signatures from the hard-to-diagnose rashes to those of confirmed cases of atopic dermatitis and psoriasis. They wanted to see if the signatures could help to reach clearer diagnoses.
The signatures revealed common immunological features as well as underlying differences. Importantly, the researchers found that the signatures allowed them to move forward and classify the indeterminate rashes. The rashes also responded to biologic therapies corresponding to the individuals’ new diagnoses.
Already, the work has identified molecules that help to define major classes of human inflammatory skin diseases. The team has also developed computer tools to help classify rashes in many other cases where the diagnosis is otherwise uncertain.
In fact, the researchers have launched a pioneering website called RashX. It is enabling practicing dermatologists and researchers around the world to submit their single-cell RNA data from their difficult cases. Such analyses are now being done at a small, but growing, number of academic medical centers.
While precision medicine for skin rashes has a long way to go yet before reaching most clinics, the UCSF team is working diligently to ensure its arrival as soon as scientifically possible. Indeed, their new data represent the beginnings of an openly available inflammatory skin disease resource. They ultimately hope to generate a standardized framework to link molecular features to disease prognosis and drug response based on data collected from clinical centers worldwide. It’s a major effort, but one that promises to improve the diagnosis and treatment of many more unusual and long-lasting rashes, both now and into the future.
Reference:
[1] Classification of human chronic inflammatory skin disease based on single-cell immune profiling. Liu Y, Wang H, Taylor M, Cook C, Martínez-Berdeja A, North JP, Harirchian P, Hailer AA, Zhao Z, Ghadially R, Ricardo-Gonzalez RR, Grekin RC, Mauro TM, Kim E, Choi J, Purdom E, Cho RJ, Cheng JB. Sci Immunol. 2022 Apr 15;7(70):eabl9165. {Epub ahead of publication]
Links:
The Promise of Precision Medicine (NIH)
Atopic Dermatitis (National Institute of Arthritis and Musculoskeletal and Skin Diseases /NIH)
Psoriasis (NIAMS/NIH)
RashX (University of California, San Francisco)
Raymond Cho (UCSF)
Jeffrey Cheng (UCSF)
NIH Support: National Institute of Arthritis and Musculoskeletal and Skin Diseases; National Center for Advancing Translational Sciences
Posted In: News
Tags: atopic dermatitis, biological therapies, biologics, computational biology, dermatology, diagnostics, eczema, genomics, immunology, inflammatory skin diseases, molecular signature, precision medicine, psoriasis, psoriasis vulgaris, rash, RashX, single-cell analysis, skin, skin rash, T cells
Artificial Intelligence Getting Smarter! Innovations from the Vision Field
Posted on February 8th, 2022 by Michael F. Chiang, M.D., National Eye Institute

One of many health risks premature infants face is retinopathy of prematurity (ROP), a leading cause of childhood blindness worldwide. ROP causes abnormal blood vessel growth in the light-sensing eye tissue called the retina. Left untreated, ROP can lead to lead to scarring, retinal detachment, and blindness. It’s the disease that caused singer and songwriter Stevie Wonder to lose his vision.
Now, effective treatments are available—if the disease is diagnosed early and accurately. Advancements in neonatal care have led to the survival of extremely premature infants, who are at highest risk for severe ROP. Despite major advancements in diagnosis and treatment, tragically, about 600 infants in the U.S. still go blind each year from ROP. This disease is difficult to diagnose and manage, even for the most experienced ophthalmologists. And the challenges are much worse in remote corners of the world that have limited access to ophthalmic and neonatal care.

Caption: Image of a neonatal retina prior to AI processing. Left: Image of a premature infant retina showing signs of severe ROP with large, twisted blood vessels; Right: Normal neonatal retina by comparison. Credit: Casey Eye Institute, Oregon Health and Science University, Portland, and National Eye Institute, NIH
Artificial intelligence (AI) is helping bridge these gaps. Prior to my tenure as National Eye Institute (NEI) director, I helped develop a system called i-ROP Deep Learning (i-ROP DL), which automates the identification of ROP. In essence, we trained a computer to identify subtle abnormalities in retinal blood vessels from thousands of images of premature infant retinas. Strikingly, the i-ROP DL artificial intelligence system outperformed even international ROP experts [1]. This has enormous potential to improve the quality and delivery of eye care to premature infants worldwide.
Of course, the promise of medical artificial intelligence extends far beyond ROP. In 2018, the FDA approved the first autonomous AI-based diagnostic tool in any field of medicine [2]. Called IDx-DR, the system streamlines screening for diabetic retinopathy (DR), and its results require no interpretation by a doctor. DR occurs when blood vessels in the retina grow irregularly, bleed, and potentially cause blindness. About 34 million people in the U.S. have diabetes, and each is at risk for DR.
As with ROP, early diagnosis and intervention is crucial to preventing vision loss to DR. The American Diabetes Association recommends people with diabetes see an eye care provider annually to have their retinas examined for signs of DR. Yet fewer than 50 percent of Americans with diabetes receive these annual eye exams.
The IDx-DR system was conceived by Michael Abramoff, an ophthalmologist and AI expert at the University of Iowa, Iowa City. With NEI funding, Abramoff used deep learning to design a system for use in a primary-care medical setting. A technician with minimal ophthalmology training can use the IDx-DR system to scan a patient’s retinas and get results indicating whether a patient should be sent to an eye specialist for follow-up evaluation or to return for another scan in 12 months.

Caption: The IDx-DR is the first FDA-approved system for diagnostic screening of diabetic retinopathy. It’s designed to be used in a primary care setting. Results determine whether a patient needs immediate follow-up. Credit: Digital Diagnostics, Coralville, IA.
Many other methodological innovations in AI have occurred in ophthalmology. That’s because imaging is so crucial to disease diagnosis and clinical outcome data are so readily available. As a result, AI-based diagnostic systems are in development for many other eye diseases, including cataract, age-related macular degeneration (AMD), and glaucoma.
Rapid advances in AI are occurring in other medical fields, such as radiology, cardiology, and dermatology. But disease diagnosis is just one of many applications for AI. Neurobiologists are using AI to answer questions about retinal and brain circuitry, disease modeling, microsurgical devices, and drug discovery.
If it sounds too good to be true, it may be. There’s a lot of work that remains to be done. Significant challenges to AI utilization in science and medicine persist. For example, researchers from the University of Washington, Seattle, last year tested seven AI-based screening algorithms that were designed to detect DR. They found under real-world conditions that only one outperformed human screeners [3]. A key problem is these AI algorithms need to be trained with more diverse images and data, including a wider range of races, ethnicities, and populations—as well as different types of cameras.
How do we address these gaps in knowledge? We’ll need larger datasets, a collaborative culture of sharing data and software libraries, broader validation studies, and algorithms to address health inequities and to avoid bias. The NIH Common Fund’s Bridge to Artificial Intelligence (Bridge2AI) project and NIH’s Artificial Intelligence/Machine Learning Consortium to Advance Health Equity and Researcher Diversity (AIM-AHEAD) Program project will be major steps toward addressing those gaps.
So, yes—AI is getting smarter. But harnessing its full power will rely on scientists and clinicians getting smarter, too.
References:
[1] Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. Brown JM, Campbell JP, Beers A, Chang K, Ostmo S, Chan RVP, Dy J, Erdogmus D, Ioannidis S, Kalpathy-Cramer J, Chiang MF; Imaging and Informatics in Retinopathy of Prematurity (i-ROP) Research Consortium. JAMA Ophthalmol. 2018 Jul 1;136(7):803-810.
[2] FDA permits marketing of artificial intelligence-based device to detect certain diabetes-related eye problems. Food and Drug Administration. April 11, 2018.
[3] Multicenter, head-to-head, real-world validation study of seven automated artificial intelligence diabetic retinopathy screening systems. Lee AY, Yanagihara RT, Lee CS, Blazes M, Jung HC, Chee YE, Gencarella MD, Gee H, Maa AY, Cockerham GC, Lynch M, Boyko EJ. Diabetes Care. 2021 May;44(5):1168-1175.
Links:
Retinopathy of Prematurity (National Eye Institute/NIH)
Diabetic Eye Disease (NEI)
Michael Abramoff (University of Iowa, Iowa City)
Bridge to Artificial Intelligence (Common Fund/NIH)
[Note: Acting NIH Director Lawrence Tabak has asked the heads of NIH’s institutes and centers to contribute occasional guest posts to the blog as a way to highlight some of the cool science that they support and conduct. This is the second in the series of NIH institute and center guest posts that will run until a new permanent NIH director is in place.]
Posted In: Generic
Tags: AI, AIM-AHEAD, AMD, artificial intelligence, blindness, brain, Bridge to Artificial Intelligence, Bridge2AI, cataract, diabetes, diabetic eye disease, diabetic retinopathy, diagnostics, eye, glaucoma, i-ROP Deep Learning, i-ROP-DL, IDx-DR, National Eye Institute, NEI, neonatal care, neural networks, ophthalmology, premature infants, retina, retinal detachment, retinopathy of prematurity, rop, Stevie Wonder, vision, vision research
A Race-Free Approach to Diagnosing Chronic Kidney Disease
Posted on October 21st, 2021 by Dr. Francis Collins

Credit: True Touch Lifestyle; crystal light/Shutterstock
Race has a long and tortured history in America. Though great strides have been made through the work of leaders like Dr. Martin Luther King, Jr. to build an equal and just society for all, we still have more work to do, as race continues to factor into American life where it shouldn’t. A medical case in point is a common diagnostic tool for chronic kidney disease (CKD), a condition that affects one in seven American adults and causes a gradual weakening of the kidneys that, for some, will lead to renal failure.
The diagnostic tool is a medical algorithm called estimated glomerular filtration rate (eGFR). It involves getting a blood test that measures how well the kidneys filter out a common waste product from the blood and adding in other personal factors to score how well a person’s kidneys are working. Among those factors is whether a person is Black. However, race is a complicated construct that incorporates components that go well beyond biological and genetic factors to social and cultural issues. The concern is that by lumping together Black people, the algorithm lacks diagnostic precision for individuals and could contribute to racial disparities in healthcare delivery—or even runs the risk of reifying race in a way that suggests more biological significance than it deserves.
That’s why I was pleased recently to see the results of two NIH-supported studies published in The New England Journal of Medicine that suggest a way to take race out of the kidney disease equation [1, 2]. The approach involves a new equation that swaps out one blood test for another and doesn’t ask about race.
For a variety of reasons, including socioeconomic issues and access to healthcare, CKD disproportionately affects the Black community. In fact, Blacks with the condition are also almost four times more likely than whites to develop kidney failure. That’s why Blacks with CKD must visit their doctors regularly to monitor their kidney function, and often that visit involves eGFR.
The blood test used in eGFR measures creatinine, a waste product produced from muscle. For about the past 20 years, a few points have been automatically added to the score of African Americans, based on data showing that adults who identify as Black, on average, have a higher baseline level of circulating creatinine. But adjusting the score upward toward normal function runs the risk of making the kidneys seem a bit healthier than they really are and delaying life-preserving dialysis or getting on a transplant list.
A team led by Chi-yuan Hsu, University of California, San Francisco, took a closer look at the current eGFR calculations. The researchers used long-term data from the Chronic Renal Insufficiency Cohort (CRIC) Study, an NIH-supported prospective, observational study of nearly 4,000 racially and ethnically diverse patients with CKD in the U.S. The study design specified that about 40 percent of its participants should identify as Black.
To look for race-free ways to measure kidney function, the researchers randomly selected more than 1,400 of the study’s participants to undergo a procedure that allows kidney function to be measured directly instead of being estimated based on blood tests. The goal was to develop an accurate approach to estimating GFR, the rate of fluid flow through the kidneys, from blood test results that didn’t rely on race.
Their studies showed that simply omitting race from the equation would underestimate GFR in Black study participants. The best solution, they found, was to calculate eGFR based on cystatin C, a small protein that the kidneys filter from the blood, in place of the standard creatinine. Estimation of GFR using cystatin C generated similarly accurate results but without the need to factor in race.
The second NIH-supported study led by Lesley Inker, Tufts Medical Center, Boston, MA, came to similar conclusions. They set out to develop new equations without race using data from several prior studies. They then compared the accuracy of their new eGFR equations to measured GFR in a validation set of 12 other studies, including about 4,000 participants.
Their findings show that currently used equations that include race, sex, and age overestimated measured GFR in Black Americans. However, taking race out of the equation without other adjustments underestimated measured GFR in Black people. Equations including both creatinine and cystatin C, but omitting race, were more accurate. The new equations also led to smaller estimated differences between Black and non-Black study participants.
The hope is that these findings will build momentum toward widespread adoption of cystatin C for estimating GFR. Already, a national task force has recommended immediate implementation of a new diagnostic equation that eliminates race and called for national efforts to increase the routine and timely measurement of cystatin C [3]. This will require a sea change in the standard measurements of blood chemistries in clinical and hospital labs—where creatinine is routinely measured, but cystatin C is not. As these findings are implemented into routine clinical care, let’s hope they’ll reduce health disparities by leading to more accurate and timely diagnosis, supporting the goals of precision health and encouraging treatment of CKD for all people, regardless of their race.
References:
[1] Race, genetic ancestry, and estimating kidney function in CKD. Hsu CY, Yang W, Parikh RV, Anderson AH, Chen TK, Cohen DL, He J, Mohanty MJ, Lash JP, Mills KT, Muiru AN, Parsa A, Saunders MR, Shafi T, Townsend RR, Waikar SS, Wang J, Wolf M, Tan TC, Feldman HI, Go AS; CRIC Study Investigators. N Engl J Med. 2021 Sep 23.
[2] New creatinine- and cystatin C-based equations to estimate GFR without race. Inker LA, Eneanya ND, Coresh J, Tighiouart H, Wang D, Sang Y, Crews DC, Doria A, Estrella MM, Froissart M, Grams ME, Greene T, Grubb A, Gudnason V, Gutiérrez OM, Kalil R, Karger AB, Mauer M, Navis G, Nelson RG, Poggio ED, Rodby R, Rossing P, Rule AD, Selvin E, Seegmiller JC, Shlipak MG, Torres VE, Yang W, Ballew SH,Couture SJ, Powe NR, Levey AS; Chronic Kidney Disease Epidemiology Collaboration. N Engl J Med. 2021 Sep 23.
[3] A unifying approach for GFR estimation: recommendations of the NKF-ASN Task Force on Reassessing the Inclusion of Race in Diagnosing Kidney Disease. Delgado C, Baweja M, Crews DC, Eneanya ND, Gadegbeku CA, Inker LA, Mendu ML, Miller WG, Moxey-Mims MM, Roberts GV, St Peter WL, Warfield C, Powe NR. Am J Kidney Dis. 2021 Sep 22:S0272-6386(21)00828-3.
Links:
Chronic Kidney Disease (National Institute of Diabetes and Digestive and Kidney Diseases/NIH)
Explaining Your Kidney Test Results: A Tool for Clinical Use (NIDDK)
Chronic Renal Insufficiency Cohort Study
Chi-yuan Hsu (University of California, San Francisco)
Lesley Inker (Tufts Medical Center, Boston)
NIH Support: National Institute of Diabetes and Digestive and Kidney Diseases
Posted In: News
Tags: African American health, African Americans, blacks, chronic kidney disease, Chronic Renal Insufficiency Cohort Study, CKD, creatinine, CRIC Study, cystatin C, diagnostics, eGFR, estimated glomerular filtration rate, genetics, GFR, glomerular filtration rate, health disparities, kidney dialysis, kidney disease, kidney failure, kidney transplantation, kidneys, muscle, precision health, precision medicine, race, renal failure
Learning to Protect Communities with COVID-19 Home Testing Programs
Posted on October 7th, 2021 by Dr. Francis Collins

Credit: Say Yes! COVID Test
With most kids now back in school, parents face a new everyday concern: determining whether their child’s latest cough or sneeze might be a sign of COVID-19. If so, parents will want to keep their child at home to protect other students and staff, while also preventing the spread of the virus in their communities. And if it’s the parent who has a new cough, they also will want to know if the reason is COVID-19 before going to work or the store.
Home tests are now coming online to help concerned people make the right choice quickly. As more COVID-19 home tests enter the U.S. marketplace, research continues to help optimize their use. That’s why NIH and the Centers for Disease Control and Prevention (CDC) are teaming up in several parts of the country to provide residents age 2 and older with free home-testing kits for COVID-19. These reliable, nasal swab tests provide yes-or-no answers in about 15 minutes for parents and anyone else concerned about their possible exposure to the novel coronavirus.
The tests are part of an initiative called Say Yes! COVID Test (SYCT) that’s evaluating how best to implement home-testing programs within range of American communities, both urban and rural. The lessons learned are providing needed science-based data to help guide public health officials who are interested in implementing similar home-testing programs in communities throughout their states.
After successful eight-week pilot programs this past spring and summer in parts of North Carolina, Tennessee, and Michigan, SYCT is partnering this fall with four new communities. They are Fulton County, GA; Honolulu County, HI; Louisville Metro, KY; and Marion County, IN.
The Georgia and Hawaii partnerships, launched on September 20, are already off to a flying start. In Fulton County, home to Atlanta and several small cities, 21,673 direct-to-consumer orders (173,384 tests) have already been received. In Honolulu County, demand for the tests has exceeded all expectations, with 91,000 orders received in the first week (728,000 tests). The online ordering has now closed in Hawaii, and the remaining tests will be distributed on the ground through the local public health department.
SYCT offers the Quidel QuickVue® At-Home COVID-19 test, which is supplied through the NIH Rapid Acceleration of Diagnostics (RADx) initiative. The antigen test uses a self-collected nasal swab sample that is placed in a test tube containing solution, followed by a test strip. Colored lines that appear on the test strip indicate a positive or negative result—similar to a pregnancy test.
The program allows residents in participating counties to order free home tests online or for in-person pick up at designated sites in their community. Each resident can ask for eight rapid tests, which equals two weekly tests over four weeks. An easy-to-navigate website like this one and a digital app, developed by initiative partner CareEvolution, are available for residents to order their tests, sign-up for testing reminders, and allow voluntary test result reporting to the public health department.
SYCT will generate data to answer several important questions about self or home-testing. They include questions about consumer demand, ensuring full community access, testing behavior, willingness to report test results, and, above all, effectiveness in controlling the spread of SARS-CoV-2, the coronavirus that causes COVID-19
Researchers at the University of North Carolina-Chapel Hill; Duke University, Durham, NC; and the UMass Chan Medical School, Worcester, MA, will help crunch the data and look for guiding themes. They will also conduct a study pre- and post-intervention to evaluate levels of SARS-CoV-2 in the community, including using measures of virus in wastewater. In addition, researchers will compare their results to other counties similar in size and infection rates, but that are not participating in a free testing initiative.
The NIH and CDC are exploring ways to scale a SYCT-like program nationally to communities experiencing surges in COVID-19. The Biden Administration also recently invoked the Defense Production Act to purchase millions of COVID-19 home tests to help accelerate their availability and offer them at a lower cost to more Americans. That encompasses many different types of people, including concerned parents who need a quick-and-accurate answer on whether their children’s cough or sneeze is COVID-19.
Links:
COVID-19 Research (NIH)
Rapid Acceleration of Diagnostics (RADx) (NIH)
NIH Support: National Institute of Biomedical Imaging and Bioengineering; National Heart, Lung, and Blood Institute; National Institute on Minority Health and Health Disparities
Posted In: News
Tags: CDC, coronavirus, COVID-19, COVID-19 testing, diagnostics, Fulton County, Georgia, Hawaii, health disparities, home tests, Honolulu County, Michigan, North Carolina, novel coronavirus, pandemic, Quidel, RADx, SARS-CoV-2, Say Yes! COVID Test, tennessee, testing, wastewater
Charting a Rapid Course Toward Better COVID-19 Tests and Treatments
Posted on August 6th, 2020 by Dr. Francis Collins
Credit: Quidel; iStock/xavierarnau
It is becoming apparent that our country is entering a new and troubling phase of the pandemic as SARS-CoV-2, the novel coronavirus that causes COVID-19, continues to spread across many states and reaches into both urban and rural communities. This growing community spread is hard to track because up to 40 percent of infected people seem to have no symptoms. They can pass the virus quickly and unsuspectingly to friends and family members who might be more vulnerable to becoming seriously ill. That’s why we should all be wearing masks when we go out of the house—none of us can be sure we’re not that asymptomatic carrier of the virus.
This new phase makes fast, accessible, affordable diagnostic testing a critical first step in helping people and communities. In recognition of this need, NIH’s Rapid Acceleration of Diagnostics (RADx) initiative, just initiated in late April, has issued an urgent call to the nation’s inventors and innovators to develop fast, easy-to-use tests for SARS-CoV-2, the novel coronavirus that causes COVID-19. It brought a tremendous response, and NIH selected about 100 of the best concepts for an intense one-week “shark-tank” technology evaluation process.
Moving ahead at an unprecedented pace, NIH last week announced the first RADx projects to come through the deep dive with flying colors and enter the scale-up process necessary to provide additional rapid testing capacity to the U.S. public. As part of the RADx initiative, seven biomedical technology companies will receive a total of $248.7 million in federal stimulus funding to accelerate their efforts to scale up new lab-based and point-of-care technologies.
Four of these projects will aim to bolster the nation’s lab-based COVID-19 diagnostics capacity by tens of thousands of tests per day as soon as September and by millions by the end of the year. The other three will expand point-of-care testing for COVID-19, making results more rapidly and readily available in doctor’s offices, urgent care clinics, long-term care facilities, schools, child care centers, or even at home.
This is only a start, and we expect that more RADx projects will advance in the coming months and begin scaling up for wide-scale use. In the meantime, here’s an overview of the first seven projects developed through the initiative, which NIH is carrying out in partnership with the Office of the Assistant Secretary of Health, the Biomedical Advanced Research and Development Authority, and the Department of Defense:
Point-of-Care Testing Approaches
Mesa Biotech. Hand-held testing device detects the genetic material of SARS-CoV-2. Results are read from a removable, single-use cartridge in 30 minutes.
Quidel. Test kit detects protein (viral antigen) from SARS-CoV-2. Electronic analyzers provide results within 15 minutes. The U.S. Department of Health and Human Service has identified this technology for possible use in nursing homes.
Talis Biomedical. Compact testing instrument uses a multiplexed cartridge to detect the genetic material of SARS-CoV-2 through isothermal amplification. Optical detection system delivers results in under 30 minutes.
Lab-based Testing Approaches
Ginkgo Bioworks. Automated system uses next-generation sequencing to scan patient samples for SARS-CoV-2’s genetic material. This system will be scaled up to make it possible to process tens of thousands of tests simultaneously and deliver results within one to two days. The company’s goal is to scale up to 50,000 tests per day in September and 100,000 per day by the end of 2020.
Helix OpCo. By combining bulk shipping of test kits and patient samples, automation, and next-generation sequencing of genetic material, the company’s goal is to process up to 50,000 samples per day by the end of September and 100,000 per day by the end of 2020.
Fluidigm. Microfluidics platform with the capacity to process thousands of polymerase chain reaction (PCR) tests for SARS-CoV-2 genetic material per day. The company’s goal is to scale up this platform and deploy advanced integrated fluidic chips to provide tens to hundreds of thousands of new tests per day in the fall of 2020. Most tests will use saliva.
Mammoth Biosciences. System uses innovative CRISPR gene-editing technology to detect key pieces of SARS-CoV-2 genetic material in patient samples. The company’s goal is to provide a multi-fold increase in testing capacity in commercial laboratories.
At the same time, on the treatment front, significant strides continue to be made by a remarkable public-private partnership called Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV). Since its formation in May, the partnership, which involves 20 biopharmaceutical companies, academic experts, and multiple federal agencies, has evaluated hundreds of therapeutic agents with potential application for COVID-19 and prioritized the most promising candidates.
Among the most exciting approaches are monoclonal antibodies (mAbs), which are biologic drugs derived from neutralizing antibodies isolated from people who’ve survived COVID-19. This week, the partnership launched two trials (one for COVID-19 inpatients, the other for COVID-19 outpatients) of a mAB called LY-CoV555, which was developed by Eli Lilly and Company, Indianapolis, IN. It was discovered by Lilly’s development partner AbCellera Biologics Inc. Vancouver, Canada, in collaboration with the NIH’s National Institute of Allergy and Infectious Diseases (NIAID). In addition to the support from ACTIV, both of the newly launched studies also receive support for Operation Warp Speed, the government’s multi-agency effort against COVID-19.
LY-CoV555 was derived from the immune cells of one of the very first survivors of COVID-19 in the United States. It targets the spike protein on the surface of SARS-CoV-2, blocking it from attaching to human cells.
The first trial, which will look at both the safety and efficacy of the mAb for treating COVID-19, will involve about 300 individuals with mild to moderate COVID-19 who are hospitalized at facilities that are part of existing clinical trial networks. These volunteers will receive either an intravenous infusion of LY-CoV555 or a placebo solution. Five days later, their condition will be evaluated. If the initial data indicate that LY-CoV555 is safe and effective, the trial will transition immediately—and seamlessly—to enrolling an additional 700 participants with COVID-19, including some who are severely ill.
The second trial, which will evaluate how LY-CoV555 affects the early course of COVID-19, will involve 220 individuals with mild to moderate COVID-19 who don’t need to be hospitalized. In this study, participants will randomly receive either an intravenous infusion of LY-CoV555 or a placebo solution, and will be carefully monitored over the next 28 days. If the data indicate that LY-CoV555 is safe and shortens the course of COVID-19, the trial will then enroll an additional 1,780 outpatient volunteers and transition to a study that will more broadly evaluate its effectiveness.
Both trials are later expected to expand to include other experimental therapies under the same master study protocol. Master protocols allow coordinated and efficient evaluation of multiple investigational agents at multiple sites as the agents become available. These protocols are designed with a flexible, rapidly responsive framework to identify interventions that work, while reducing administrative burden and cost.
In addition, Lilly this week started a separate large-scale safety and efficacy trial to see if LY-CoV555 can be used to prevent COVID-19 in high-risk residents and staff at long-term care facilities. The study isn’t part of ACTIV.
NIH-funded researchers have been extremely busy over the past seven months, pursuing every avenue we can to detect, treat, and, ultimately, end this devasting pandemic. Far more work remains to be done, but as RADx and ACTIV exemplify, we’re making rapid progress through collaboration and a strong, sustained investment in scientific innovation.
Links:
Coronavirus (COVID-19) (NIH)
Rapid Acceleration of Diagnostics (RADx)
Video: NIH RADx Delivering New COVID-19 Testing Technologies to Meet U.S. Demand (YouTube)
Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV)
Explaining Operation Warp Speed (U.S. Department of Health and Human Resources/Washington, D.C.)
“NIH delivering new COVID-19 testing technologies to meet U.S. demand,” NIH news release,” July 31, 2020.
“NIH launches clinical trial to test antibody treatment in hospitalized COVID-19 patients,” NIH new release, August 4, 2020.
“NIH clinical trial to test antibodies and other experimental therapeutics for mild and moderate COVID-19,” NIH news release, August 4, 2020.
Posted In: News
Tags: AbCellerra Biologics, Accelerating COVID-19 Therapeutic Interventions and Vaccines, ACTIV, antibodies, coronavirus, COVID-19, COVID-19 testing, COVID-19 treatment, diagnostics, Eli Lilly and Company, Fluidigm, Gingko Bioworks, Helix OpCo, lab-based testing, LY-CoV555, mAbs, Mammoth Biosciences, master protocols, Mesa Biotech, monoclonal antibody, novel coronavirus, Operation Warp Speed, pandemic, point-of-care tests, Quidel, RADx, Rapid Acceleration of Diagnostics Initiative, saliva test, SARS-CoV-2, spike protein, Talis Biomedical, therapeutics
Racing to Develop Fast, Affordable, Accessible Tests for COVID-19
Posted on July 23rd, 2020 by Dr. Francis Collins

Credit: iStock/peshkov
Developing faster, more convenient ways of testing for coronavirus disease 2019 (COVID-19) will be essential to our efforts to end this deadly pandemic. Despite the tremendous strides that have been made in diagnostics over the past seven months, we still need more innovation.
We need reliable, affordable tests for the presence SARS-CoV-2—the novel coronavirus that causes COVID-19—that do not take hours or days to deliver results. We need tests that are more user friendly, and that don’t rely on samples collected by swabs that have to be inserted deep into the nose by someone wearing PPE. We need tests that can be performed at the point-of-care, whether a doctor’s office, urgent care clinic, long-term care facility, or even a home. Ideally, such tests should also be able to integrate with mobile devices to convey results and transmit data seamlessly. Above all, we need tests that are accessible to everyone.
Most current diagnostic tests for SARS-CoV-2 involve detecting viral genetic material using a decades-old technology called the polymerase chain reaction (PCR). If there’s even a tiny bit of viral genetic material in a patient’s sample, PCR can amplify the material millions of times so that it can be readily detected. The problem is that this amplification process is time-consuming and requires a thermal cycling machine that’s generally operated by trained personnel in sophisticated lab settings.
To spur the creation of new approaches that can rapidly expand access to testing, NIH launched the Rapid Acceleration of Diagnostics (RADx) program in late April 2020. This fast-paced, innovative effort, conducted in partnership with the Office of the Assistant Secretary of Health, the Biomedical Advanced Research and Development Authority (BARDA), and the Department of Defense, is supported by $1.5 billion in federal stimulus funding. The goal? To expand diagnostic testing capacity for COVID-19 in the United States to about 6 million tests per day by December. That’s quite a leap forward because our nation’s current testing capacity is currently about 1 million tests per day.
Just yesterday, I joined other NIH leaders in authoring a special report in the New England Journal of Medicine that describes RADx’s main activities, and provides an update on the remarkable progress that’s been made in just three short months [1]. In a nutshell, RADx consists of four components: RADx-tech, RADx Advanced Technology Platforms (RADx-ATP). RADx Radical (RADx-rad), and RADx Underserved Populations (RADx-UP).
Though all parts of RADx are operating on a fast-track, RADx-tech has embraced its rapid timelines in a can-do manner unlike anything that I’ve encountered in my 27 years in government. Here’s how the process, which has been likened to a scientific “shark tank,” works.
Once an applicant submits a test idea to RADx-tech, it’s reviewed within a day by a panel of 30 experts. If approved, the application moves to a highly competitive “shark-tank” in which a team of experts spend about 150 to 200 person-hours with the applicant evaluating the technical, clinical, and commercial strengths and weaknesses of the proposed test.
From there, a detailed proposal is presented to a steering committee, and then sent to NIH. If we at NIH think it’s a great idea, promising early-stage technologies enter what’s called “phase one” development, with considerable financial support and the expectation that the applicant will hit its validation milestones within a month. Technologies that succeed can then go to “phase two”, where support is provided for scale-up of tests for meeting regulatory requirements and supporting manufacture, scale-up, and distribution.
The major focus of RADx-tech is to simplify and speed diagnostic testing for COVID-19. Tests now under development include a variety of mobile devices that can be used at a doctor’s office or other point-of-care settings, and give results in less than an hour. In addition, about half of the tests now under development use saliva or another alternative to samples gathered via nasal swabs.
As Americans think about how to move back safely into schools, workspaces, and other public areas in the era of COVID-19, it is clear that we need to figure out ways to make it easier for everyone to get tested. To attain that goal, RADx has three other components that build on different aspects of this social imperative:
• RADx Advanced Technology Platforms (RADx-ATP). This program offers a rapid-response application process for firms with existing point-of-care technologies authorized by the Food and Drug Administration (FDA) for detecting SARS-CoV-2. These technologies are already advanced enough that they don’t need the shark tank. The RADx-ATP program provides support for scaling up production to between 20,000 and 100,000 tests per day by the fall. Another component of this program provides support for expanding automated “mega-labs” to increase testing capacity across the country by another 100,000 to 250,000 tests per day.
• RADx Radical (RADx-rad). The program seeks to fuel the development of truly futuristic testing technologies. For example, it supports projects that use biomarkers to detect an infection or predict the severity of disease, including the likelihood of developing COVID-related multisystem inflammatory syndrome in children (MIS-C). Other areas of interest include the use of biosensors to detect the presence of the virus in a person’s breath and the analysis of wastewater to conduct community-based surveillance.
• RADx Underserved Populations (RADx-UP). Data collected over the past several months make it clear that Blacks, Latinxs, and American Indians/Alaska Natives are hospitalized and die of COVID-19 at disproportionately higher rates than other groups. RADx-UP aims to engage underserved communities to improve access to testing. Such actions will include closely examining the factors that have led to the disproportionate burden of the pandemic on underserved populations, as well as building infrastructure that can be leveraged to provide optimal access and uptake of SARS-CoV-2 testing in such communities.
At NIH, we have great hopes for what RADx-supported research will do to help bring to an end the greatest public health crisis of our generation. Yet the benefits may not end there. The diagnostic testing technologies developed here will have many other applications moving forward. Long after the COVID-19 pandemic becomes a chapter in history books, I’m convinced the RADx model of rapid innovation will be inspiring future generations of researchers as they look for creative new ways to address other diseases and conditions.
Reference:
[1] Rapid scaling up of COVID-19 diagnostic testing in the United States—The NIH RADx Initiative. Tromberg BJ, Schwetz TA, Perez-Stable E, Hodes RJ. Woychick RP, Bright RA, Fleurence RL, Collins FS. NEJM; 2020 July 16. [Online publication ahead of print]
Links:
Coronavirus (COVID-19) (NIH)
Rapid Acceleration of Diagnostics (RADx)
“NIH mobilizes national innovation initiative for COVID-19 diagnostics,” NIH news release, April 29, 2020.
Posted In: News
Tags: Alaskan Native, American Indian, BARDA, bioengineering, black, coronavirus, COVID-19, COVID-19 testing, Department of Defense, diagnostic testing, diagnostics, Latinix, novel coronavirus, pandemic, point-of-care tests, RADx, RADx-ATP, RADx-rad, RADx-tech, RADx-UP, Rapid Acceleration of Diagnostics Initiative, saliva test, SARS-CoV-2, Shark Tank, technology, tests
Pop-Up Testing Lab Shows Volunteer Spirit Against Deadly Pandemic
Posted on May 12th, 2020 by Dr. Francis Collins

Caption: Postdoc Jenny Hamilton volunteered to work on coronavirus testing at the Innovative Genomics Institute. Behind her is one of the lab’s liquid-handling systems, which robotically extracts RNA from patient samples before another machine can detect whether that RNA comes from the coronavirus. Credit: Max & Jules Photography.
On March 19, 2020, California became the first U. S. state to issue a stay-at-home order to halt the spread of SARS-CoV-2, the novel coronavirus that causes COVID-19. The order shuttered research labs around the state, and thousands of scientists began sheltering at home and shifting their daily focus to writing papers and grants, analyzing data from past experiments, and catching up on their scientific reading.
That wasn’t the case for everyone. Some considered the order as presenting a perfect opportunity to volunteer, sometimes outside of their fields of expertise, to help their state and communities respond to the pandemic.
One of those willing to pitch in is Jennifer Doudna, University of California, Berkeley (UC Berkeley) and executive director of the school’s Innovative Genomics Institute (IGI), a partnership with the University of California, San Francisco (UC San Francisco). She is also recognized as a pioneer in the development of the popular gene-editing technology called CRISPR.
Doudna, an NIH-supported structural biochemist with no experience in virology or clinical diagnostics, decided that she and her IGI colleagues could establish a pop-up testing lab at their facility. Their job: boost the SARS-CoV-2 testing capacity in her community.
It was a great idea, but a difficult one to execute. The first daunting step was acquiring Clinical Laboratory Improvement Amendments (CLIA) certification. This U. S. certification ensures that quality standards are met for laboratory testing of human blood, body fluid, and other specimens for medical purposes. CLIA certification is required not only to perform such testing in the IGI lab space, but for Doudna’s graduate students, postdocs, and volunteers to process patient samples.
Still, fate was on their side. Doudna and her team partnered with UC Berkeley’s University Health Services to extend the student health center’s existing CLIA certification to the IGI space. And because of the urgency of the pandemic, federal review of the extension request was expedited and granted in a few weeks.
The next challenge was technological. Doudna’s team had to make sure that its diagnostic system was as good or better than those of other SARS-CoV-2 testing platforms. With great care and attention to lab safety, the team began assembling two parallel workstreams: one a semi-manual method to get going right away and the other a faster, automated, robotic method to transition to when ready.
Soon, patient samples began arriving in the lab to be tested for the presence of genetic material (RNA) from SARS-CoV-2, an indication that a person is infected with the virus. The diagnostic system was also soon humming along, with Doudna’s automated workstream having the capacity to process 384 samples in parallel.
The pop-up lab—known formally as the IGI SARS-CoV-2 Diagnostic Testing Laboratory—is funded through philanthropy and staffed by more than 50 volunteers from IGI, UC Berkeley, UC San Francisco, and local data-management companies. Starting on April 6, the lab was fully operational, capable of running hundreds of tests daily with a 24-hour turnaround time for results. A positive test requires that at least two out of three SARS-CoV-2 genomic targets return a positive signal, and the method uses de-identified barcoded sample data to protect patient privacy.
Doudna intends to keep the pop-up lab open as long as her community needs it. So far, they’ve provided testing to UC Berkeley students and staff, first responders (including the entire Berkeley Fire Department), and several members of the city’s homeless population. She says that availability of samples will soon be the rate-limiting step in their sample-analysis pipeline and hopes continued partnerships with local health officials will enable them to work at full capacity to deliver thousands of test results rapidly.
Doudna says she’s been amazed by the team spirit of her lab members and other local colleagues who have come together around a crisis. They’ve gotten the job done by contributing their different skills and resources, including behind-the-scenes efforts by the university’s leadership and staff, philanthropists, city officials, and state government workers.
Although Doudna and her team intend to publish their work to help others follow suit [1], she says the experience has also provided her with many intangible rewards. It has highlighted the value of resilience and adaptation, as well as given her a newfound appreciation for the complexity and precision of operations in the commercial clinical labs that are a routine part of our medical care.
Although the COVID-19 pandemic seems to have thrust all of us into a time warp, in which weeks sometimes feel like months, there is much to do. The amount of work needed to tame this virus is significant and requires an all-hands-on-deck mentality, which NIH and the biomedical research community have embraced fully.
Doudna is not alone. Other labs around the country are engaged in similar efforts. At the NIH’s main campus in Bethesda, MD, staff at the clinical laboratory in the Clinical Center rapidly set up testing for SARS-CoV-2 RNA, and have now tested more than 1,000 NIH staff. Researchers at the Broad Institute of MIT and Harvard partnered with the city of Cambridge, MA, to pilot COVID-19 surveillance in homeless shelters and skilled nursing and assisted living facilities located there.
Hats off to everyone who goes the extra mile to get us through this tough time. I am so gratified when, guided by compassion and dogged determination of the human spirit, science leads the way and provides much needed hope for our future.
Reference:
[1] Blueprint for a Pop-up SARS-CoV-2 Testing Lab. Innovative Genomics Institute SARS-CoV-2 Testing Consortium, Hockemeyer D, Fyodor U, Doudna JA. 2020. medRxiv. Preprint posted on April 12, 2020.
Links:
Coronavirus (COVID-19) (NIH)
CLIA Law & Regulations (Centers for Disease Control and Prevention)
Innovative Genomic Institute (Berkeley, CA)
Doudna Lab (University of California, Berkeley)
Posted In: Generic
Tags: Broad Institute, California, Cambridge MA, CLIA, coronavirus, COVID-19, COVID-19 testing, CRISPR, diagnostics, IGI SARS-CoV-2 Diagnostic Testing Laboratory, Innovative Genomics Institute, NIH Clinical Center, novel coronavirus, pandemic, pop-up lab, SARS-CoV-2, UC Berkeley, volunteerism
Study Finds Nearly Everyone Who Recovers From COVID-19 Makes Coronavirus Antibodies
Posted on May 7th, 2020 by Dr. Francis Collins

Credit: NIH
There’s been a lot of excitement about the potential of antibody-based blood tests, also known as serology tests, to help contain the coronavirus disease 2019 (COVID-19) pandemic. There’s also an awareness that more research is needed to determine when—or even if—people infected with SARS-CoV-2, the novel coronavirus that causes COVID-19, produce antibodies that may protect them from re-infection.
A recent study in Nature Medicine brings much-needed clarity, along with renewed enthusiasm, to efforts to develop and implement widescale antibody testing for SARS-CoV-2 [1]. Antibodies are blood proteins produced by the immune system to fight foreign invaders like viruses, and may help to ward off future attacks by those same invaders.
In their study of blood drawn from 285 people hospitalized with severe COVID-19, researchers in China, led by Ai-Long Huang, Chongqing Medical University, found that all had developed SARS-CoV-2 specific antibodies within two to three weeks of their first symptoms. Although more follow-up work is needed to determine just how protective these antibodies are and for how long, these findings suggest that the immune systems of people who survive COVID-19 have been be primed to recognize SARS-CoV-2 and possibly thwart a second infection.
Specifically, the researchers determined that nearly all of the 285 patients studied produced a type of antibody called IgM, which is the first antibody that the body makes when fighting an infection. Though only about 40 percent produced IgM in the first week after onset of COVID-19, that number increased steadily to almost 95 percent two weeks later. All of these patients also produced a type of antibody called IgG. While IgG often appears a little later after acute infection, it has the potential to confer sustained immunity.
To confirm their results, the researchers turned to another group of 69 people diagnosed with COVID-19. The researchers collected blood samples from each person upon admission to the hospital and every three days thereafter until discharge. The team found that, with the exception of one woman and her daughter, the patients produced specific antibodies against SARS-CoV-2 within 20 days of their first symptoms of COVID-19.
Meanwhile, innovative efforts are being made on the federal level to advance COVID-19 testing. The NIH just launched the Rapid Acceleration of Diagnostics (RADx) Initiative to support a variety of research activities aimed at improving detection of the virus. As I recently highlighted on this blog, one key component of RADx is a “shark tank”-like competition to encourage science and engineering’s most inventive minds to develop rapid, easy-to-use technologies to test for the presence of SARS-CoV-2.
On the serology testing side, the NIH’s National Cancer Institute has been checking out kits that are designed to detect antibodies to SARS-CoV-2 and have found mixed results. In response, the Food and Drug Administration just issued its updated policy on antibody tests for COVID-19. This guidance sets forth precise standards for laboratories and commercial manufacturers that will help to speed the availability of high-quality antibody tests, which in turn will expand the capacity for rapid and widespread testing in the United States.
Finally, it’s important to keep in mind that there are two different types of SARS-CoV-2 tests. Those that test for the presence of viral nucleic acid or protein are used to identify people who are acutely infected and should be immediately quarantined. Tests for IgM and/or IgG antibodies to the virus, if well-validated, indicate a person has previously been infected with COVID-19 and is now potentially immune. Two very different types of tests—two very different meanings.
There’s still a way to go with both virus and antibody testing for COVID-19. But as this study and others begin to piece together the complex puzzle of antibody-mediated immunity, it will be possible to learn more about the human body’s response to SARS-CoV-2 and home in on our goal of achieving safe, effective, and sustained protection against this devastating disease.
Reference:
[1] Antibody responses to SARS-CoV-2 in patients with COVID-19. Long QX, Huang AI, et al. Nat Med. 2020 Apr 29. [Epub ahead of print]
Links:
[Coronaviruses](http://Coronavirus %28COVID-19%29 %28NIH%29 https://www.nih.gov/health-information/coronavirus) (NIH)
“NIH Begins Study to Quantify Undetected Cases of Coronavirus Infection,” NIH News Release, April 10, 2020.
“NIH mobilizes national innovation initiative for COVID-19 diagnostics,” NIH News Release, April 29, 2020.
Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (Revised), May 2020 (Food and Drug Administration)
Posted In: News
Tags: antibodies, antibody testing, China, COVID-19, COVID-19 antibody test, COVID-19 testing, diagnostics, FDA, IgG, IgM, novel coronavirus, pandemic, Polivcy for Coronavirus Disease-2019 Tests During the Public Health Emergency, RADx, Rapid Acceleration of Diagnostics Initiative, SARS-CoV-2, serology testing, testing