imaging – NIH Director's Blog (original) (raw)
AI Model Takes New Approach to Performing Diagnostic Tasks in Multiple Cancer Types
Posted on October 3rd, 2024 by Dr. Monica M. Bertagnolli

Credit: Donny Bliss/NIH, Adobe Stock
In recent years, medical researchers have been looking for ways to use artificial intelligence (AI) technology for diagnosing cancer. So far, most AI models have been developed to perform specific tasks in cancer diagnosis, such as detecting cancer presence or predicting a tumor’s genetic profile in certain cancer types. But what if an AI system could be more flexible, like a large language model such as ChatGPT, performing a variety of diagnostic tasks across multiple cancer types?
As reported in the journal Nature, researchers have developed an AI system that can perform a wide range of cancer evaluation tasks and outperforms current AI methods in tasks like cancer cell detection and tumor origin identification. It was tested on 19 cancer types, leading the researchers to refer to it as “ChatGPT-like” in its flexibility. According to the research team, whose work is supported in part by NIH, this is also the first AI model based on analyzing slide images to not only accurately predict if a cancer is likely to respond to treatment, but also to validate these predictions across multiple patient groups around the world.
Today, when doctors order a biopsy to find out if cancer is present, those samples are sent to a pathologist, who examines the tissues or cells under a microscope to determine if they are cancerous. The team behind this AI model, led by Kun-Hsing Yu, Harvard Medical School, Boston, recognized that pathologists must analyze a wide variety of disease samples. To make accurate diagnoses in different cancer types, they must take many subtle factors into account.
Most earlier attempts to devise an AI model to analyze tissue samples have depended on training computers to recognize one cancer type at a time. In the new work, the researchers developed a more general-purpose pathology AI system that could analyze a broader range of tissues and sample types. To develop their Clinical Histopathology Imaging Evaluation Foundation (CHIEF) model, the researchers used an AI approach known as self-supervised learning. In this method, a computer is given large volumes of data, in this case 15 million pathology images, to allow it to identify intrinsic patterns and structures. This process allows a computer to “learn” from experience to identify informative features in a vast data set.
The tool was then trained further on more than 60,500 whole-slide images in tissues collected from 19 different parts of the body—such as the lungs, breast, prostate, kidney, brain, and bladder—to bolster the model’s ability to capture similarities and differences among cancer types. This training data was in part comprised of data from The Cancer Genome Atlas (TCGA) program and the Genotype-Tissue Expression (GTEx) Project, both NIH-supported resources. The researchers directed the model to consider both the image as a whole and its finer details, enabling it to interpret the image in a broader context than one region. They then put CHIEF to the test, using another 19,491 whole-slide images from 32 independent slide sets collected from 24 hospitals around the world.
They found that CHIEF worked equally well no matter how the samples were collected (biopsy or surgical excision) and in different clinical settings. In addition to detecting cancers and predicting a cancer’s tissue of origin, CHIEF also predicted with 70% accuracy whether a tissue carried one among dozens of genetic mutations that are commonly seen in cancers. CHIEF showed an ability to predict whether a sample contained mutated copies of 18 genes that oncologists use to make treatment decisions. CHIEF could predict better than earlier models how long a patient was likely to survive following a cancer diagnosis and how aggressively a particular cancer would grow.
This is all good news, but there’s much more work ahead before an AI model like this could be used in the clinic. Next steps for the researchers include training the model on images of tissues from rare cancers, as well as from pre-cancerous and non-cancerous conditions. With continued development and validation, the researchers aim to enable the system to identify cancers most likely to benefit from targeted or experimental therapies in hopes of improving outcomes for more people with cancer in diverse clinical settings around the world.
Reference:
Wang X, et al. A pathology foundation model for cancer diagnosis and prognosis prediction. Nature. DOI: 10.1038/s41586-024-07894-z (2024).
NIH Support: National Institute of General Medical Sciences, National Cancer Institute
Posted In: Health, Science, technology
Tags: AI, artificial intelligence, cancer, cancer diagnosis, computer learning, genetic mutations, genetics, imaging, pathology, technology
Mapping Psilocybin’s Brain Effects to Explore Potential for Treating Mental Health Disorders
Posted on August 15th, 2024 by Dr. Monica M. Bertagnolli

Credit: Donny Bliss/NIH
Psilocybin is a natural ingredient found in “magic mushrooms.” A single dose of this psychedelic can distort a person’s perception of time and space, as well as their sense of self, for hours. It can also trigger strong emotions, ranging from euphoria to fear. While psilocybin comes with health risks and isn’t recommended for recreational use, there’s growing evidence that—under the right conditions—its effects on the brain might be harnessed in the future to help treat substance use disorders or mental illnesses.
To explore this potential, it will be important to understand how psilocybin exerts its effects on the human brain. Now, a study in Nature supported in part by NIH has taken a step in this direction, using functional brain mapping in healthy adults before, during, and after taking psilocybin to visualize its impact. While earlier studies in animals suggested that psilocybin makes key brain areas more adaptable or “plastic,” this new research aims to clarify changes in the function of larger brain networks and their connection to the experiences people have with this psychedelic drug.
In the study, the team led by Joshua Siegel, Nico Dosenbach and Ginger Nicol, Washington University School of Medicine in St. Louis, recruited seven healthy adults to take, in separate sessions, a high dose of psilocybin and the stimulant methylphenidate, the generic form of Ritalin, under controlled conditions. Because taking psilocybin comes with a risk for having disturbing or negative experiences, a pair of trained experts stayed with each participant throughout the sessions to provide guidance and support. They also helped participants prepare for and process the experience afterwards.
To visually capture the impact of psilocybin on the brain, the researchers had each participant undergo an average of 18 functional MRI brain scan visits. Four of the study’s participants also returned six to 12 months later to take an additional psilocybin dose. Comparisons of the brain images revealed profound and widespread, but temporary, changes to the brain’s functional networks. While an individual’s functional brain network is typically as distinctive as a fingerprint, psilocybin made the participants’ brain networks look so similar in the scans that the researchers couldn’t tell them apart. In addition, by following the brain scans with specialized questionnaires given to elicit details about participants’ subjective experiences with psilocybin, the researchers were able to generate precise data on each person’s unique impressions and associated changes in their brain networks.
For all the participants, psilocybin desynchronized the brain’s default mode network, an interconnected set of brain areas that are most active when people are daydreaming or otherwise not engaged in any focused, goal-directed mental activity. By comparison, the default mode network remained stable after study participants took methylphenidate. Once the effects of psilocybin wore off, brain function returned almost to its original state. However, the researchers did note small but potentially important differences in each participant’s brain scans after taking psilocybin that remained for weeks.
The researchers suggest that the short-term changes in the default mode network likely explain psilocybin’s psychedelic effects, including the way it changes the way a person thinks about themselves in relation to other people and the world. They also suggest that the more subtle, longer-term effects they observed might indicate that the brain is more flexible in the weeks following a dose of psilocybin in ways that could allow for a healthier state. This may help explain preliminary research showing that psilocybin may have benefits for treating substance use disorders, as well as depression, anxiety, and other mental health conditions.
Though these findings are encouraging, they should not be seen as a reason to try psilocybin without clinician supervision or use it to self-medicate. The drug is not proven or approved as a treatment for any condition, and its unsupervised use comes with serious risks. The researchers hope that with much more clinical study of how and why this drug affects individuals in the powerful ways that it does, this kind of research may one day lead to a greater understanding of the human brain and promising new interventions that improve mental health.
Reference:
Siegel JS, et al. Psilocybin desynchronizes the human brain. Nature. DOI: 10.1038/s41586-024-07624-5 (2024).
NIH Support: National Institute of Mental Health, National Institute on Drug Abuse, National Institute of Neurological Disorders and Stroke
Tags: anxiety, brain, default mode network, depression, imaging, mental health disorders, neuroscience, psilocybin, psychedelic, substance use disorders
Most Detailed 3D Reconstruction of Human Brain Tissue Ever Produced Yields Surprising Insights
Posted on May 30th, 2024 by Dr. Monica M. Bertagnolli
Researchers have developed a detailed 3D reconstruction of a cubic millimeter of brain tissue. Credit: Images in video from Google Research & Lichtman Lab, Harvard University. Renderings by D. Berger, Harvard. Video compiled by Donny Bliss/NIH
The NIH _Brain Research Through Advancing Innovative Neurotechnologies_® (BRAIN) Initiative has expanded scientists’ understanding of the human brain in recent years, offering fascinating insights into the ways that individual cells and complex neural circuits interact dynamically to enable us to think, feel, and act. But neuroscientists still have much more to learn about how our brains are put together at the most fundamental, subcellular level.
As a step in that direction, in a new study supported in part by the NIH BRAIN Initiative and reported in the journal Science, researchers have created the most detailed nanoscale resolution map ever produced of a cubic millimeter of brain tissue, about the size of half a grain of rice.
Despite its small size, this fragment of healthy brain contained about 57,000 cells of various types, 230 millimeters of blood vessels, 150 million neural connections, or synapses, and the protective myelin that insulates neurons. To capture it all in vivid detail, the researchers relied on electron microscopy to amass an impressive 1,400 terabytes of imaging data. For perspective, one terabyte of data is enough to store 100,000 photos on your smartphone.
While there are many more details yet to analyze given the sheer quantity of data, this impressively detailed subcellular map has already revealed multiple brain structures that have never been seen before. This includes a class of triangular neurons in deep brain layers being described for the first time. The map also revealed axons, the long extensions of nerve cells that carry electrical impulses, with as many as 50 synapses and other unusual structures, including axons arranged into extensive spiraling patterns that now warrant further study.
The findings come from a team led by Jeff W. Lichtman, Harvard University, Cambridge, MA, and Viren Jain, Google Research, Mountain View, CA. They recognized that fully understanding the human brain requires knowledge of its most basic construction. While the imaging technologies needed to produce this kind of map were available, there were other barriers, including a limited availability of healthy and high-quality human brain tissue samples for study.
Most biopsies of the brain are done to examine or take out abnormal growths of cells or tissues, making them unsuitable for understanding the normal makeup of the brain. In this case, the researchers were able to obtain a tiny sample from the brain tissue removed and destined for disposal during the normal course of surgery for a patient with epilepsy. The researchers first stained the preserved sample to make the cells easier to trace individually before slicing it into 5,000 thin layers for microscopic imaging.
To put those slices back together into a complete 3D reconstruction, the researchers relied on artificial intelligence (AI) models. Because the dataset is too large for any one group to fully analyze, they’ve made it all freely available to the research community in an online resource. They’ve also provided tools for its further analysis and proofreading.
While there is plenty still left to uncover, the findings offer proof-of-principle that it’s possible to visualize the brain at this very detailed level. This is crucial groundwork for new research now supported by the BRAIN Initiative Connectivity Across Scales (BRAIN CONNECTS) program. BRAIN CONNECTS will develop and scale up tools to produce an equally detailed map of a complete mouse brain, which is about 1,000 times larger than the human brain fragment. The researchers now hope their 3D map and others like it will be put to work to understand both normal and disordered brain function more fully.
Reference:
[1] Shapson-Coe A, et al. A petavoxel fragment of human cerebral cortex reconstructed at nanoscale resolution. Science. DOI: 10.1126/science.adk4858 (2024).
NIH Support: NIH BRAIN Initiative, National Institute of Mental Health
Posted In: News, Science, technology
Tags: 3d reconstruction, brain, BRAIN Initiative, brain tissue, data, imaging, neural circuitry, neuroscience, research tools
The Amazing Brain: Turning Conventional Wisdom on Brain Anatomy on its Head
Posted on November 28th, 2023 by John Ngai, PhD, NIH BRAIN Initiative
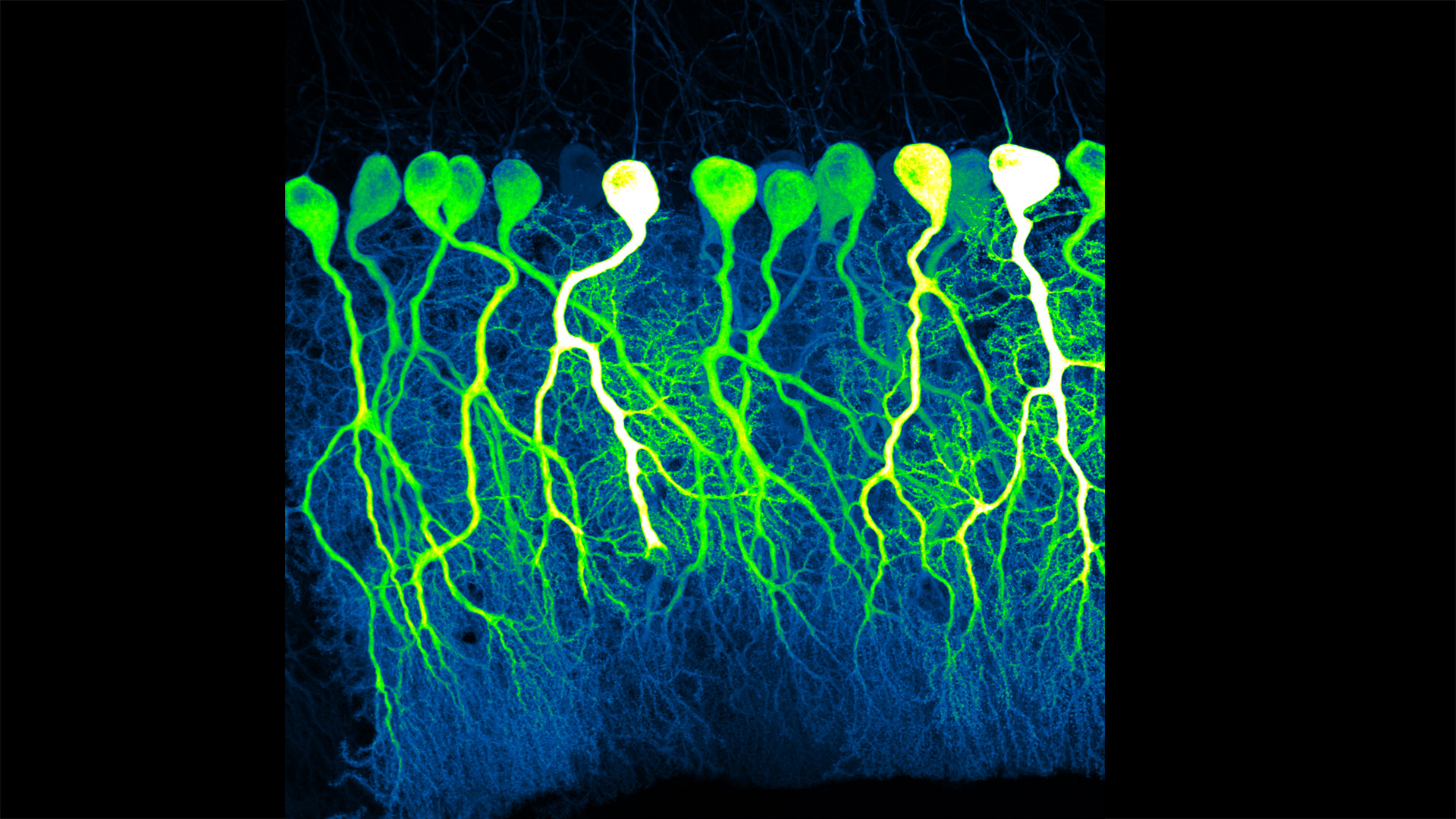
Credit: Silas Busch, The University of Chicago
Silas Busch at the University of Chicago captured this slightly eerie scene, noting it reminded him of people shuffling through the dark of night. What you’re really seeing are some of the largest neurons in the mammalian brain, known as Purkinje cells. The photo won first place this year in the _Brain Research through Advancing Innovative Neurotechnologies_® (BRAIN) Initiative’s annual Show Us Your BRAINs! Photo and Video Contest.
While humans have them, too, the Purkinje cells pictured here are in the brain of a mouse. The head-like shapes you see in the image are the so-called soma, or the neurons’ cell bodies. Extending downwards are the heavily branched dendrites, which act like large antennae, receiving thousands of inputs from the rest of the body.
One reason this picture is such a standout, explains Busch, is because of what you don’t see. You’ll notice that only a few cells are fluorescently labeled and therefore lit up in green, leaving the rest in shadows. As a result, it’s possible to trace the detailed branches of individual Purkinje cells and make out their intricate forms. As it turns out, this ability to trace Purkinje cells so precisely led Busch, who is a graduate student in Christian Hansel’s lab focused on the neurobiology of learning and memory, to a surprising discovery, which the pair recently reported in Science.1
Purkinje cells connect to nerve fibers that “climb up” from the brain stem, which connects your brain and spinal cord to help control breathing, heart rate, balance and more. Scientists thought that each Purkinje cell received only one of these climbing fibers from the brain stem on its single primary branch.
However, after carefully tracing thousands of Purkinje cells in brain tissue from both mice and humans, the researchers have now shown that Purkinje cells and climbing fibers don’t always have a simple one-to-one relationship. In fact, Busch and Hansel found more than 95 percent of human Purkinje cells have multiple primary branches, not just one. In mice, that ratio is closer to 50 percent.
The researchers went on to show that mouse Purkinje cells with multiple primary branches often also receive multiple climbing fibers. The discovery rewrites the textbook idea of how Purkinje cells in the brain and climbing fibers from the brainstem are anatomically arranged.
Not surprisingly, those extra connections in the cerebellum (located in the back of the brain) also have important functional implications. When Purkinje cells have just one climbing fiber input, the new study shows, the whole cell receives each signal equally and responds accordingly. But, in cells with multiple climbing fiber inputs, the researchers could detect differences across a single cell depending on which primary branch received an input.
What this means is that Purkinje cells in the brain have much more computational power than had been appreciated. That extra brain power has important implications for understanding how brain circuits can adapt and respond to changes outside the body that now warrant further study. The new findings may have implications also for understanding the role of these Purkinje cell connections in some neurological and developmental disorders, including autism2 and a movement disorder known as cerebellar ataxia.
As they say, a picture is worth a thousand words. And this winning image comes as a reminder that we still have more to learn from careful study of basic brain anatomy, with important implications for human health and disease.
References:
[1] SE Busch and C Hansel. Climbing fiber multi-innervation of mouse Purkinje dendrites with arborization common to human. Science. DOI: 10.1126/science.adi1024. (2023).
[2] DH Simmons et al. Sensory Over-responsivity and Aberrant Plasticity in Cerebellar Cortex in a Mouse Model of Syndromic Autism. Biological Psychiatry: Global Open Science. DOI: 10.1016/j.bpsgos.2021.09.004. (2021).
Can Bioprinted Skin Substitutes Replace Traditional Grafts for Treating Burn Injuries and Other Serious Skin Wounds?
Posted on October 17th, 2023 by Lawrence Tabak, D.D.S., Ph.D.
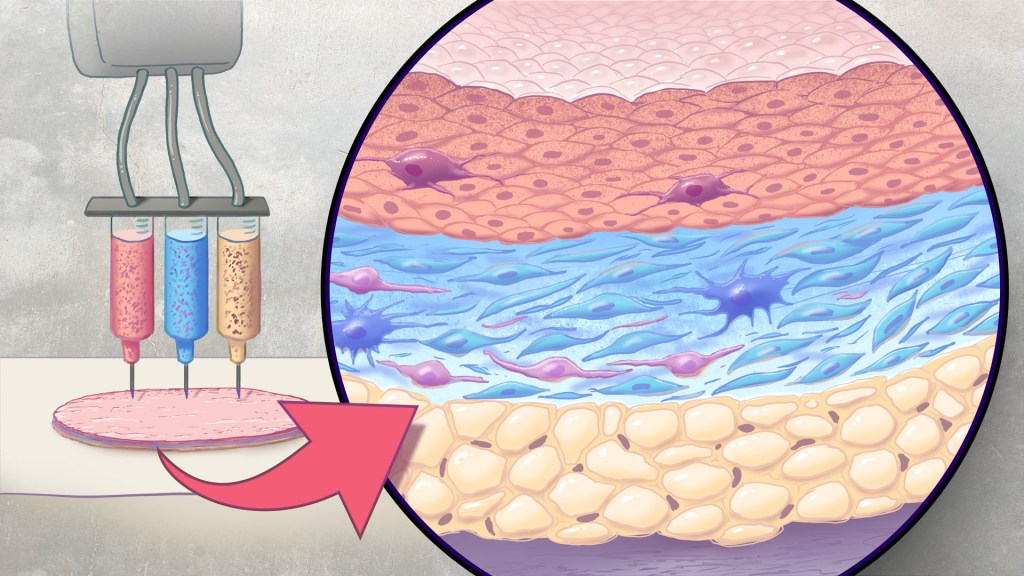
Artificial skin is printed by layering specific cell mixtures to mimic human skin: epidermis (top), dermis (middle) and hypodermis (bottom). Credit: Donny Bliss/NIH
Each year in the U.S., more than 500,000 people receive treatment for burn injuries and other serious skin wounds.1 To close the most severe wounds with less scarring, doctors often must surgically remove skin from one part of a person’s body and use it to patch the injured site. However, this is an intensive process, and some burn patients with extensive skin loss do not have sufficient skin available for grafting. Scientists have been exploring ways to repair these serious skin wounds without skin graft surgery.
An NIH-funded team recently showed that bioprinted skin substitutes may serve as a promising alternative to traditional skin grafts in preclinical studies reported in _Science Translational Medicine._2 The approach involves a portable skin bioprinter system that deposits multiple layers of skin directly into a wound. The recent findings add to evidence that bioprinting technology can successfully regenerate human-like skin to allow healing. While this approach has yet to be tested in people, it confirms that such technologies already can produce skin constructs with the complex structures and multiple cell types present in healthy human skin.
This latest work comes from a team led by Adam Jorgensen and Anthony Atala at Wake Forest School of Medicine’s Wake Forest Institute for Regenerative Medicine, Winston-Salem, NC. Members of the Atala lab and their colleagues had earlier shown it was possible to isolate two major skin cell types found in the skin’s outer (epidermis) and middle (dermis) layers from a small biopsy of healthy skin, expand the number of cells in the lab and then deliver the cells directly into an injury using a specially designed bioprinter.3 Using integrated imaging technology to scan a wound, computer software “prints” cells right into an injury, mimicking two of our skin’s three natural layers.
In the new study, Atala’s team has gone even further to construct skin substitutes that mimic the structure of human skin and that include six primary human skin cell types. They then used their bioprinter to produce skin constructs with all three layers found in healthy human skin: epidermis, dermis, and hypodermis.
To put their skin substitutes to the test, they first transplanted them into mice. Their studies showed that the bioprinted skin encouraged the rapid growth of new blood vessels and had other features of normal-looking, healthy skin. The researchers were able to confirm that their bioprinted skin implants successfully integrated into the animals’ regenerated skin to speed healing.
Studies in a pig model of wound healing added to evidence that such bioprinted implants can successfully repair full-thickness wounds, meaning those that extend through all three layers of skin. The bioprinted skin patches allowed for improved wound healing with less scarring. They also found that the bioprinted grafts encouraged activity in the skin from genes known to play important roles in wound healing.
It’s not yet clear if this approach will work as well in the clinic as it does in the lab. To make it feasible, the researchers note there’s a need for improved approaches to isolating and expanding the needed skin cell types. Nevertheless, these advances come as encouraging evidence that bioprinted skin substitutes could one day offer a promising alternative to traditional skin grafts with the capacity to help even more people with severe burns or other wounds.
References:
[1] Burn Incidence Fact Sheet. American Burn Association
[2] AM Jorgensen, et al. Multicellular bioprinted skin facilitates human-like skin architecture in vivo. Science Translational Medicine DOI: 10.1126/scitranslmed.adf7547 (2023).
[3] M Albanna, et al. In Situ Bioprinting of Autologous Skin Cells Accelerates Wound Healing of Extensive Excisional Full-Thickness Wounds. Scientific Reports DOI: 10.1038/s41598-018-38366-w (2019).
NIH Support: National Institute of Arthritis and Musculoskeletal and Skin Diseases
Posted In: Health, News, Science, technology
Tags: bioprinting, burn, burn treatment, dermis, epidermis, hypodermis, imaging, skin, skin cell, skin graft surgery, skin grafts, wound healing, wounds
How to Feed a Macrophage
Posted on August 22nd, 2023 by Lawrence Tabak, D.D.S., Ph.D.
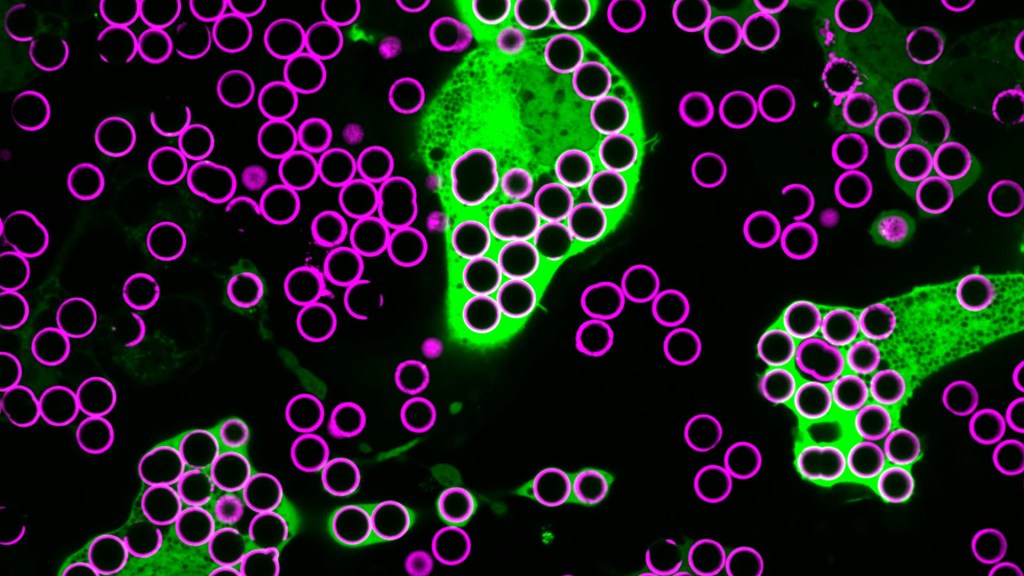
Credit: Annalise Bond, Morrissey Lab, University of California, Santa Barbara
For Annalise Bond, a graduate student in the lab of Meghan Morrissey, University of California, Santa Barbara (UCSB), macrophages are “the professional eaters of our immune system.” Every minute of every day, macrophages somewhere in the body are gorging themselves to remove the cellular debris that builds up in our tissues and organs.
In this image, Bond caught several macrophages (green) doing what they do best: shoveling it in—in this case, during a lab experiment. The macrophages are consuming silica beads (purple) prepared with biochemicals that whet their appetites. Each bead measures about five microns in diameter. That’s roughly the size of a bacterium or a spent red blood cell—debris that a macrophage routinely consumes.
When Bond snapped this image, she noticed a pattern that reminded her of a childhood tabletop game called Hungry Hungry Hippos. Kids press a lever attached to the mouth of a plastic hippo, its lower jaw flaps open, and the challenge is to fill the mouth with as many marbles as possible . . . just like the macrophages eating beads.
Bond adjusted the colors in the photo to make them pop. She then entered it into UCSB’s 2023 Art of Science contest with the caption of Hungry Hungry Macrophages, winning high marks for drawing the association.
Though the caption was written in fun, Bond studies in earnest a fascinating biological question: How do macrophages know what to eat in the body and what to leave untouched?
In her studies, Bond coats the silica beads shown above with a lipid bilayer to mimic a cell membrane. To that coating, she adds various small molecules and proteins as “eat-me” signals often found on the surface of dying cells. Some of the signals are well characterized; but many aren’t, meaning there’s still a lot to learn about what makes a macrophage “particularly hungry” and what makes a particular target cell “extra tasty.”
Capturing fluorescent images of macrophages under the microscope, Bond counts up how many beads are eaten. Beads bearing no signal to stimulate their appetite might get eaten occasionally. But when an especially enticing signal is added, macrophages will gorge themselves until they can’t eat anymore.
In the experiment pictured above, the beads contain the antibody immunoglobulin G (IgG), which tags foreign pathogens for macrophage removal. Interestingly, IgG antibody responses also play an important role in cancer immunotherapies, in which the immune system is unleashed to fight cancer.
Among its many areas of study, the NIH-supported Morrissey lab’s wants to understand better how macrophages interact with cancer cells. They want to learn how to make cancer cells even tastier to macrophages and program their elimination from the body. Sorting out the signals will be challenging, but we know that macrophages will take a bite at the right ones. They are, after all, professional eaters.
Links:
Cancer Immunotherapy (NIH)
Annalise Bond (University of California, Santa Barbara)
Morrissey Lab (University of California, Santa Barbara)
Art of Science (University of California, Santa Barbara)
NIH Support: National Institute of General Medical Sciences
3D Animation Captures Viral Infection in Action
Posted on August 1st, 2023 by Lawrence Tabak, D.D.S., Ph.D.
With the summer holiday season now in full swing, the blog will also swing into its annual August series. For most of the month, I will share with you just a small sampling of the colorful videos and snapshots of life captured in a select few of the hundreds of NIH-supported research labs around the country.
To get us started, let’s turn to the study of viruses. Researchers now can generate vast amounts of data relatively quickly on a virus of interest. But data are often displayed as numbers or two-dimensional digital images on a computer screen. For most virologists, it’s extremely helpful to see a virus and its data streaming in three dimensions. To do so, they turn to a technological tool that we all know so well: animation.
This research animation features the chikungunya virus, a sometimes debilitating, mosquito-borne pathogen transmitted mainly in developing countries in Africa, Asia and the Americas. The animation illustrates large amounts of research data to show how the chikungunya virus infects our cells and uses its specialized machinery to release its genetic material into the cell and seed future infections. Let’s take a look.
In the opening seconds, you see how receptor binding glycoproteins (light blue), which are proteins with a carbohydrate attached on the viral surface, dock with protein receptors (yellow) on a host cell. At five seconds, the virus is drawn inside the cell. The change in the color of the chikungunya particle shows that it’s coated in a vesicle, which helps the virus make its way unhindered through the cytoplasm.
At 10 seconds, the virus then enters an endosome, ubiquitous bubble-like compartments that transport material from outside the cell into the cytosol, the fluid part of the cytoplasm. Once inside the endosome, the acidic environment makes other glycoproteins (red, blue, yellow) on the viral surface change shape and become more flexible and dynamic. These glycoproteins serve as machinery that enables them to reach out and grab onto the surrounding endosome membrane, which ultimately will be fused with the virus’s own membrane.
As more of those fusion glycoproteins grab on, fold back on themselves, and form into hairpin-like shapes, they pull the membranes together. The animation illustrates not only the changes in protein organization, but the resulting effects on the integrity of the membrane structures as this dynamic process proceeds. At 53 seconds, the viral protein shell, or capsid (green), which contains the virus’ genetic instructions, is released back out into the cell where it will ultimately go on to make more virus.
This remarkable animation comes from Margot Riggi and Janet Iwasa, experts in visualizing biology at the University of Utah’s Animation Lab, Salt Lake City. Their data source was researcher Kelly Lee, University of Washington, Seattle, who collaborated closely with Riggi and Iwasa on this project. The final product was considered so outstanding that it took the top prize for short videos in the 2022 BioArt Awards competition, sponsored by the Federation of American Societies for Experimental Biology (FASEB).
The Lee lab uses various research methods to understand the specific shape-shifting changes that chikungunya and other viruses perform as they invade and infect cells. One of the lab’s key visual tools is cryo-electron microscopy (Cryo-EM), specifically cryo-electron tomography (cryo-ET). Cryto-ET enables complex 3D structures, including the intermediate state of biological reactions, to be captured and imaged in remarkably fine detail.
In a study in the journal Nature Communications [1] last year, Lee’s team used cryo-ET to reveal how the chikungunya virus invades and delivers its genetic cargo into human cells to initiate a new infection. While Lee’s cryo-ET data revealed stages of the virus entry process and fine structural details of changes to the virus as it enters a cell and starts an infection, it still represented a series of snapshots with missing steps in between. So, Lee’s lab teamed up with The Animation Lab to help beautifully fill in the gaps.
Visualizing chikungunya and similar viruses in action not only makes for informative animations, it helps researchers discover better potential targets to intervene in this process. This basic research continues to make progress, and so do ongoing efforts to develop a chikungunya vaccine [2] and specific treatments that would help give millions of people relief from the aches, pains, and rashes associated with this still-untreatable infection.
References:
[1] Visualization of conformational changes and membrane remodeling leading to genome delivery by viral class-II fusion machinery. Mangala Prasad V, Blijleven JS, Smit JM, Lee KK. Nat Commun. 2022 Aug 15;13(1):4772. doi: 10.1038/s41467-022-32431-9. PMID: 35970990; PMCID: PMC9378758.
[2] Experimental chikungunya vaccine is safe and well-tolerated in early trial, National Institute of Allergy and Infectious Diseases news release, April 27, 2020.
Links:
Chikungunya Virus (Centers for Disease Control and Prevention, Atlanta)
Global Arbovirus Initiative (World Health Organization, Geneva, Switzerland)
The Animation Lab (University of Utah, Salt Lake City)
Video: Janet Iwasa (TED Speaker)
Lee Lab (University of Washington, Seattle)
BioArt Awards (Federation of American Societies for Experimental Biology, Rockville, MD)
NIH Support: National Institute of General Medical Sciences; National Institute of Allergy and Infectious Diseases
Posted In: Cool Videos
Tags: 2022 BioArt Awards, animation, chikungunya, chikungunya vaccine, cryo-electron tomography, cryo-EM, cryo-ET, cytoplasm, cytosol, endosome, FASEB, global health, glycoprotein, imaging, infection, mosquito-borne illnesses, structural biology, vesicle, virology, virus
Cryo-EM Scores Again
Posted on July 25th, 2023 by Lawrence Tabak, D.D.S., Ph.D.

Caption: Researchers recently published the near-atomic structure of the neuronal pore CALHMI. Credit: Donny Bliss, NIH
Human neurons are long, spindly structures, but if you could zoom in on their surfaces at super-high resolution, you’d see surprisingly large pores. They act as gated channels that open and close for ions and other essential molecules of life to pass in and out the cell. This rapid exchange of ions and other molecules is how neurons communicate, and why we humans can sense, think, move, and respond to the world around us [1].
Because these gated channels are so essential to neurons, mapping their precise physical structures at high-resolution has profound implications for informing future studies on the brain and nervous system. Good for us in these high-tech times that structural biologists keep getting better at imaging these 3D pores.
In fact, as just published in the journal Nature Communications [2], a team of NIH-supported scientists imaged the molecular structure of a gated pore of major research interest. The pore is called calcium homeostasis modulator 1 (CALHM1). Pictured below, you can view its 3D structure at near atomic resolution [2]. Keep in mind, this relatively large neuronal pore still measures approximately 50,000 times smaller than the width of a hair.

Caption: Human CALHM1 channel. It has an eight-protein assembly pattern. Note the different colored arm-like structures above. White dot (center) is ruthenium red, a chemical that blocks off the channel. Credit: Furkawa lab/Cryo-EM Facility/Cold Spring Harbor Laboratory, NY
The structure comes from a research team led by Hiro Furukawa, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. He and his team relied on cryo-electron microscopy (cryo-EM) to produce the first highly precise 3D models of CALHM1.
Cryo-EM involves flash-freezing molecules in liquid ethane and bombarding them with electrons to capture their images with a special camera. When all goes well, cryo-EM can reveal the structure of intricate macromolecular complexes in a matter of weeks.
Furukawa’s team had earlier studied CALHM1 from chickens with cryo-EM [3], and their latest work reveals that the human version is quite similar. Eight copies of the CALHM1 protein assemble to form the circular channel. Each of the protein subunits has a flexible arm that allows it to reach into the central opening, which the researchers now suspect allows the channels to open and close in a highly controlled manner. The researchers have likened the channels’ eight flexible arms to the arms of an octopus.
The researchers also found that fatty molecules called phospholipids play a critical role in stabilizing and regulating the eight-part channel. They used simulations to demonstrate how pockets in the CALHM1 channel binds this phospholipid over cholesterol to shore up the structure and function properly. Interestingly, these phospholipid molecules are abundant in many healthy foods, such as eggs, lean meats, and seafood.
Researchers knew that an inorganic chemical called ruthenium red can block the function of the CALHM1 channel. They’ve now shown precisely how this works. The structural details indicate that ruthenium red physically lodges in and plugs up the channel.
These details also may be useful in future efforts to develop drugs designed to target and modify the function of these channels in helpful ways. For instance, on our tongues, the channel plays a role in our ability to perceive sweet, sour, or umami (savory) flavors. In our brains, studies show the abnormal function of CALHM1 may be implicated in the plaques that accumulate in the brains of people with Alzheimer’s disease.
There are far too many other normal and abnormal functions to mention here in this brief post. Suffice it to say, I’ll look forward to seeing what this enabling research yields in the years ahead.
References:
[1] On the molecular nature of large-pore channels. Syrjanen, J., Michalski, K., Kawate, T., and Furukawa, H. J Mol Biol. 2021 Aug 20;433(17):166994. DOI: 10.1016/j.jmb.2021.166994. Epub 2021 Apr 16. PMID: 33865869; PMCID: PMC8409005.
[2] Structure of human CALHM1 reveals key locations for channel regulation and blockade by ruthenium red. Syrjänen JL, Epstein M, Gómez R, Furukawa H. Nat Commun. 2023 Jun 28;14(1):3821. DOI: 10.1038/s41467-023-39388-3. PMID: 37380652; PMCID: PMC10307800.
[3] Structure and assembly of calcium homeostasis modulator proteins. Syrjanen JL, Michalski K, Chou TH, Grant T, Rao S, Simorowski N, Tucker SJ, Grigorieff N, Furukawa H. Nat Struct Mol Biol. 2020 Feb;27(2):150-159. DOI: 10.1038/s41594-019-0369-9. Epub 2020 Jan 27. PMID: 31988524; PMCID: PMC7015811.
Links:
Brain Basics: The Life and Death of a Neuron (National Institute of Neurological Disorders and Stroke/NIH)
Alzheimer’s Disease (National Institute on Aging/NIH)
Furukawa Lab (Cold Spring Harbor Lab, Cold Spring Harbor, NY)
NIH Support: National Institute of Neurological Disorders and Stroke; National Institute of Mental Health
Posted In: News
Tags: Alzheimer’s disease, calcium homeostasis modulator 1, CALHM1, cryo-electron microscopy, cryo-EM, imaging, neuron, phospholipid, ruthenium red, structural biology, taste
Changes in Normal Brain Connections Linked to Eating Disorders
Posted on April 11th, 2023 by Lawrence Tabak, D.D.S., Ph.D.
Anyone who has ever had a bad habit knows how vexingly difficult breaking it can be. The reason is the repeated action, initially linked to some type of real or perceived reward, over time changes the way our very brains are wired to work. The bad habit becomes automatic, even when the action does us harm or we no longer wish to do it.
Now an intriguing new study shows that the same bundled nerve fibers, or brain circuits, involved in habit formation also can go awry in people with eating disorders. The findings may help to explain why eating disorders are so often resistant to will power alone. They also may help to point the way to improved approaches to treating eating disorders, suggesting strategies that adjust the actual brain circuitry in helpful ways.
These latest findings, published in the journal Science Translational Medicine, come from the NIH-supported Casey Halpern, University of Pennsylvania’s Perelman School of Medicine, Philadelphia, and Cara Bohon, Stanford University School of Medicine, Stanford, CA [1].
Halpern, Bohon, and colleagues were interested in a growing body of evidence linking habitual behaviors to mental health conditions, most notably substance use disorders and addictions. But what especially intrigued them was recent evidence also suggesting a possible role for habitual behaviors in the emergence of eating disorders.
To look deeper into the complex circuitry underlying habit formation and any changes there that might be associated with eating disorders, they took advantage of a vast collection of data from the NIH-funded Human Connectome Project (HCP). It was completed several years ago and now serves as a valuable online resource for researchers.
The HCP offers a detailed wiring map of a normal human brain. It describes all the structural and functional neural connections based on careful analyses of hundreds of high-resolution brain scans. These connections are then layered with genetic, behavioral, and other types of data. This incredible map now allows researchers to explore and sometimes uncover the roots of neurological and mental health conditions within the brain’s many trillions of connections.
In the new study, Halpern, Bohon, and colleagues did just that. First, they used sophisticated mapping methods in 178 brain scans from the HCP data to locate key portions of a brain region called the striatum, which is thought to be involved in habit formation. What they really wanted to know was whether circuits operating within the striatum were altered in some way in people with binge eating disorder or bulimia nervosa.
To find out, the researchers recruited 34 women who have an eating disorder and, with their consent, imaged their brains using a variety of techniques. Twenty-one participants were diagnosed with binge eating disorder, and 13 had bulimia nervosa. For comparison purposes, the researchers looked at the same brain circuits in 19 healthy volunteers.
The two groups were otherwise similar in terms of their ages, weights, and other features. But the researchers suspected they might find differences between the healthy group and those with an eating disorder in brain circuits known to have links to habitual behaviors. And, indeed, they did.
In comparison to a “typical” brain, those from people with an eating disorder showed striking changes in the connectivity of a portion of the striatum known as the putamen. That’s especially notable because the putamen is known for its role in learning and movement control, including reward, thinking, and addiction. What’s more, those observed changes in the brain’s connections and circuitry in this key brain area were more evident in people whose eating disorder symptoms and emotional eating were more frequent and severe.
Using other brain imaging methods in 10 of the volunteers (eight with binge eating disorder and two healthy controls), the researchers also connected those changes in the habit-forming brain circuits to high levels of a protein receptor that responds to dopamine. Dopamine is an important chemical messenger in the brain involved in pleasure, motivation, and learning. They also observed in those with eating disorders structural changes in the architecture of the densely folded, outer layer of the brain known as grey matter.
While there’s much more to learn, the researchers note the findings may lead to future treatments aimed to modify the brain circuitry in beneficial ways. Indeed, Halpern already has encouraging early results from a small NIH-funded clinical trial testing the ability of deep brain stimulation (DBS) in people with binge eating disorder to disrupt signals that drive food cravings in another portion of the brain associated with reward and motivation, known as the nucleus accumbens, [2]. In DBS, doctors implant a pacemaker-like device capable of delivering harmless therapeutic electrical impulses deep into the brain, aiming for the spot where they can reset the abnormal circuitry that’s driving eating disorders or other troubling symptoms or behaviors.
But the latest findings published in Science Translational Medicine now suggest other mapped brain circuits as potentially beneficial DBS targets for tackling binge eating, bulimia nervosa, or other life-altering, hard-to-treat eating disorders. They also may ultimately have implications for treating other conditions involving various other forms of compulsive behavior.
These findings should come as a source of hope for the family and friends of the millions of Americans—many of them young people—who struggle with eating disorders. The findings also serve as an important reminder for the rest of us that, despite common misconceptions that disordered eating is a lifestyle choice, these conditions are in fact complex and serious mental health problems driven by fundamental changes in the brain’s underlying circuitry.
Finding new and more effective ways to treat serious eating disorders and other compulsive behaviors is a must. It will require equally serious ongoing efforts to unravel their underlying causes and find ways to alter their course—and this new study is an encouraging step in that direction.
References:
[1] Human habit neural circuitry may be perturbed in eating disorders. Wang AR, Kuijper FM, Barbosa DAN, Hagan KE, Lee E, Tong E, Choi EY, McNab JA, Bohon C, Halpern CH. Sci Transl Med. 2023 Mar 29;15(689):eabo4919.
[2] Pilot study of responsive nucleus accumbens deep brain stimulation for loss-of-control eating. Shivacharan RS, Rolle CE, Barbosa DAN, Cunningham TN, Feng A, Johnson ND, Safer DL, Bohon C, Keller C, Buch VP, Parker JJ, Azagury DE, Tass PA, Bhati MT, Malenka RC, Lock JD, Halpern CH. Nat Med. 2022 Sep;28(9):1791-1796.
Links:
Eating Disorders (National Institute of Mental Health/NIH)
Casey Halpern (Penn Medicine, Philadelphia)
Cara Bohon (Stanford University, Stanford, CA)
NIH Support: National Institute of Mental Health; National Institute of Neurological Disorders and Stroke
Posted In: News
Tags: binge eating disorder, brain, brain circuits, brain imaging, bulimia nervosa, clinical trial, connectomics, DBS, deep brain stimulation, eating, eating disorders, habitual behaviors, habitual eating, HCP, Human Connectome Project, imaging, mental health, neuroanatomy, nucleus accumbens, putamen, striatum
Saving Fat for Lean Times
Posted on February 28th, 2023 by Lawrence Tabak, D.D.S., Ph.D.
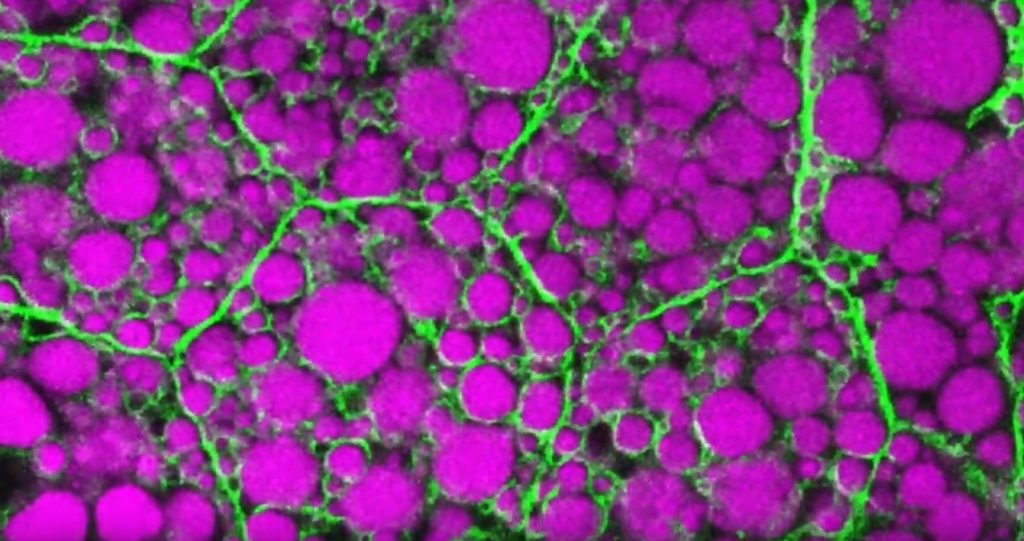
Credit: Rupali Ugrankar, Henne Lab, University of Texas Southwestern Medical Center, Dallas
Humans and all multi-celled organisms, or metazoans, have evolved through millennia into a variety of competing shapes, sizes, and survival strategies. But all metazoans still share lots of intriguing cell biology, including the ability to store excess calories as fat. In fact, many researchers now consider fat-storing cells to be “nutrient sinks,” or good places for the body to stash excess sugars and lipids. Not only can these provide energy needed to survive a future famine, this is a good way to sequester extra molecules that could prove toxic to cells and organs.
Here’s something to think about the next time you skip a meal. Fat-storing cells organize their fat reserves spatially, grouping them into specific pools of lipid types, in order to generate needed energy when food is scarce.
That’s the story behind this striking image taken in a larval fruit fly (Drosophila melanogaster). The image captures fat-storing adipocytes in an organ called a fat body, where a larval fruit fly stores extra nutrients. It’s like the fat tissue in mammals. You can see both large and small lipid droplets (magenta) inside polygon-shaped fat cells, or adipocytes, lined by their plasma membranes (green). But notice that the small lipid droplets are more visibly lined by green, as only these are destined to be saved for later and exported when needed into the fly’s bloodstream.
Working in Mike Henne’s lab at the University of Texas Southwestern Medical Center, Dallas, research associate Rupali Ugrankar discovered how this clever fat-management system works in Drosophila [1]. After either feeding flies high-or-extremely low-calorie diets, Ugrankar used a combination of high-resolution fluorescence confocal microscopy and thin-section transmission electron microscopy to provide a three-dimensional view of adipocytes and their lipid droplets inside.
She observed two distinct sizes of lipid droplets and saw that only the small ones clustered at the cell surface membrane. The adipocytes contorted their membrane inward to grab these small droplets and package them into readily exportable energy bundles.
Ugrankar saw that during times of plenty, a protein machine could fill these small membrane-wrapped fat droplets with lots of triacylglycerol, a high-energy, durable form of fat storage. Their ready access at the surface of the adipocyte allows the fly to balance lipid storage locally with energy release into its blood in times of famine.
Ugrankar’s adeptness at the microscope resulted in this beautiful photo, which was earlier featured in the American Society for Cell Biology’s Green Fluorescent Protein Image and Video Contest. But her work and that of many others help to open a vital window into nutrition science and many critical mechanistic questions about the causes of obesity, insulin resistance, hyperglycemia, and even reduced lifespan.
Such basic research will provide the basis for better therapies to correct these nutrition-related health problems. But the value of basic science must not be forgotten—some of the most important leads could come from a tiny insect in its larval state that shares many aspects of mammalian metabolism.
Reference:
[1] Drosophila Snazarus regulates a lipid droplet population at plasma membrane-droplet contacts in adipocytes. Ugrankar R, Bowerman J, Hariri H, Chandra M, et al. Dev Cell. 2019 Sep 9;50(5):557-572.e5.
Links:
The Interactive Fly (Society for Developmental Biology, Rockville, MD)
Henne Lab (University of Texas Southwestern Medical Center, Dallas)
NIH Support: National Institute of General Medical Sciences
Posted In: Snapshots of Life
Tags: 2019 Green Fluorescent Protein Image and Video Contest, adipocyte, American Society for Cell Biology, basic research, cell biology, Drosophila melanogaster, fat, fat body, fat cells, fat storage, fluorescence microscopy, fruit fly, hyperglycemia, imaging, insulin resistance, lifespan, lipid droplet, lipid storage, lipids, metabolism, model organisms, nutrient sink, obesity, triacylglycerol