influenza – NIH Director's Blog (original) (raw)
Immune Resilience is Key to a Long and Healthy Life
Posted on June 20th, 2023 by Lawrence Tabak, D.D.S., Ph.D.

Caption: A new measure of immunity called immune resilience is helping researchers find clues as to why some people remain healthier even in the face of varied inflammatory stressors. Credit: Modified from Shutterstock/Ground Picture
Do you feel as if you or perhaps your family members are constantly coming down with illnesses that drag on longer than they should? Or, maybe you’re one of those lucky people who rarely becomes ill and, if you do, recovers faster than others.
It’s clear that some people generally are more susceptible to infectious illnesses, while others manage to stay healthier or bounce back more quickly, sometimes even into old age. Why is this? A new study from an NIH-supported team has an intriguing answer [1]. The difference, they suggest, may be explained in part by a new measure of immunity they call immune resilience—the ability of the immune system to rapidly launch attacks that defend effectively against infectious invaders and respond appropriately to other types of inflammatory stressors, including aging or other health conditions, and then quickly recover, while keeping potentially damaging inflammation under wraps.
The findings in the journal Nature Communications come from an international team led by Sunil Ahuja, University of Texas Health Science Center and the Department of Veterans Affairs Center for Personalized Medicine, both in San Antonio. To understand the role of immune resilience and its effect on longevity and health outcomes, the researchers looked at multiple other studies including healthy individuals and those with a range of health conditions that challenged their immune systems.
By looking at multiple studies in varied infectious and other contexts, they hoped to find clues as to why some people remain healthier even in the face of varied inflammatory stressors, ranging from mild to more severe. But to understand how immune resilience influences health outcomes, they first needed a way to measure or grade this immune attribute.
The researchers developed two methods for measuring immune resilience. The first metric, a laboratory test called immune health grades (IHGs), is a four-tier grading system that calculates the balance between infection-fighting CD8+ and CD4+ T-cells. IHG-I denotes the best balance tracking the highest level of resilience, and IHG-IV denotes the worst balance tracking the lowest level of immune resilience. An imbalance between the levels of these T cell types is observed in many people as they age, when they get sick, and in people with autoimmune diseases and other conditions.
The researchers also developed a second metric that looks for two patterns of expression of a select set of genes. One pattern associated with survival and the other with death. The survival-associated pattern is primarily related to immune competence, or the immune system’s ability to function swiftly and restore activities that encourage disease resistance. The mortality-associated genes are closely related to inflammation, a process through which the immune system eliminates pathogens and begins the healing process but that also underlies many disease states.
Their studies have shown that high expression of the survival-associated genes and lower expression of mortality-associated genes indicate optimal immune resilience, correlating with a longer lifespan. The opposite pattern indicates poor resilience and a greater risk of premature death. When both sets of genes are either low or high at the same time, immune resilience and mortality risks are more moderate.
In the newly reported study initiated in 2014, Ahuja and his colleagues set out to assess immune resilience in a collection of about 48,500 people, with or without various acute, repetitive, or chronic challenges to their immune systems. In an earlier study, the researchers showed that this novel way to measure immune status and resilience predicted hospitalization and mortality during acute COVID-19 across a wide age spectrum [2].
The investigators have analyzed stored blood samples and publicly available data representing people, many of whom were healthy volunteers, who had enrolled in different studies conducted in Africa, Europe, and North America. Volunteers ranged in age from 9 to 103 years. They also evaluated participants in the Framingham Heart Study, a long-term effort to identify common factors and characteristics that contribute to cardiovascular disease.
To examine people with a wide range of health challenges and associated stresses on their immune systems, the team also included participants who had influenza or COVID-19, and people living with HIV. They also included kidney transplant recipients, people with lifestyle factors that put them at high risk for sexually transmitted infections, and people who’d had sepsis, a condition in which the body has an extreme and life-threatening response following an infection.
The question in all these contexts was the same: How well did the two metrics of immune resilience predict an individual’s health outcomes and lifespan? The short answer is that immune resilience, longevity, and better health outcomes tracked together well. Those with metrics indicating optimal immune resilience generally had better health outcomes and lived longer than those who had lower scores on the immunity grading scale. Indeed, those with optimal immune resilience were more likely to:
- Live longer,
- Resist HIV infection or the progression from HIV to AIDS,
- Resist symptomatic influenza,
- Resist a recurrence of skin cancer after a kidney transplant,
- Survive COVID-19, and
- Survive sepsis.
The study also revealed other interesting findings. While immune resilience generally declines with age, some people maintain higher levels of immune resilience as they get older for reasons that aren’t yet known, according to the researchers. Some people also maintain higher levels of immune resilience despite the presence of inflammatory stress to their immune systems such as during HIV infection or acute COVID-19. People of all ages can show high or low immune resilience. The study also found that higher immune resilience is more common in females than it is in males.
The findings suggest that there is a lot more to learn about why people differ in their ability to preserve optimal immune resilience. With further research, it may be possible to develop treatments or other methods to encourage or restore immune resilience as a way of improving general health, according to the study team.
The researchers suggest it’s possible that one day checkups of a person’s immune resilience could help us to understand and predict an individual’s health status and risk for a wide range of health conditions. It could also help to identify those individuals who may be at a higher risk of poor outcomes when they do get sick and may need more aggressive treatment. Researchers may also consider immune resilience when designing vaccine clinical trials.
A more thorough understanding of immune resilience and discovery of ways to improve it may help to address important health disparities linked to differences in race, ethnicity, geography, and other factors. We know that healthy eating, exercising, and taking precautions to avoid getting sick foster good health and longevity; in the future, perhaps we’ll also consider how our immune resilience measures up and take steps to achieve or maintain a healthier, more balanced, immunity status.
References:
[1] Immune resilience despite inflammatory stress promotes longevity and favorable health outcomes including resistance to infection. Ahuja SK, Manoharan MS, Lee GC, McKinnon LR, Meunier JA, Steri M, Harper N, Fiorillo E, Smith AM, Restrepo MI, Branum AP, Bottomley MJ, Orrù V, Jimenez F, Carrillo A, Pandranki L, Winter CA, Winter LA, Gaitan AA, Moreira AG, Walter EA, Silvestri G, King CL, Zheng YT, Zheng HY, Kimani J, Blake Ball T, Plummer FA, Fowke KR, Harden PN, Wood KJ, Ferris MT, Lund JM, Heise MT, Garrett N, Canady KR, Abdool Karim SS, Little SJ, Gianella S, Smith DM, Letendre S, Richman DD, Cucca F, Trinh H, Sanchez-Reilly S, Hecht JM, Cadena Zuluaga JA, Anzueto A, Pugh JA; South Texas Veterans Health Care System COVID-19 team; Agan BK, Root-Bernstein R, Clark RA, Okulicz JF, He W. Nat Commun. 2023 Jun 13;14(1):3286. doi: 10.1038/s41467-023-38238-6. PMID: 37311745.
[2] Immunologic resilience and COVID-19 survival advantage. Lee GC, Restrepo MI, Harper N, Manoharan MS, Smith AM, Meunier JA, Sanchez-Reilly S, Ehsan A, Branum AP, Winter C, Winter L, Jimenez F, Pandranki L, Carrillo A, Perez GL, Anzueto A, Trinh H, Lee M, Hecht JM, Martinez-Vargas C, Sehgal RT, Cadena J, Walter EA, Oakman K, Benavides R, Pugh JA; South Texas Veterans Health Care System COVID-19 Team; Letendre S, Steri M, Orrù V, Fiorillo E, Cucca F, Moreira AG, Zhang N, Leadbetter E, Agan BK, Richman DD, He W, Clark RA, Okulicz JF, Ahuja SK. J Allergy Clin Immunol. 2021 Nov;148(5):1176-1191. doi: 10.1016/j.jaci.2021.08.021. Epub 2021 Sep 8. PMID: 34508765; PMCID: PMC8425719.
Links:
COVID-19 Research (NIH)
HIV Info (NIH)
Sepsis (National Institute of General Medical Sciences/NIH)
Sunil Ahuja (University of Texas Health Science Center, San Antonio)
Framingham Heart Study (National Heart, Lung, and Blood Institute/NIH)
“A Secret to Health and Long Life? Immune Resilience, NIAID Grantees Report,” NIAID Now Blog, June 13, 2023
NIH Support: National Institute of Allergy and Infectious Diseases; National Institute on Aging; National Institute of Mental Health; National Institute of General Medical Sciences; National Heart, Lung, and Blood Institute
Posted In: News
Tags: aging, cardiovascular disease, CD4+, CD8+, COVID-19, Framingham Heart Study, genomics, health disparities, HIV, IHG, immune health grades, immune resilience, immune system, immunity, influenza, kidney transplantation, lifespan, longevity, sepsis, skin cancer, T cells
Experimental mRNA Vaccine May Protect Against All 20 Influenza Virus Subtypes
Posted on December 6th, 2022 by Lawrence Tabak, D.D.S., Ph.D.

Caption: Messenger RNA (mRNA)– nanoparticle vaccine encoding hemagglutinin antigens (H with number) from all 20 known influenza subtypes.
Flu season is now upon us, and protecting yourself and loved ones is still as easy as heading to the nearest pharmacy for your annual flu shot. These vaccines are formulated each year to protect against up to four circulating strains of influenza virus, and they generally do a good job of this. What they can’t do is prevent future outbreaks of more novel flu viruses that occasionally spill over from other species into humans, thereby avoiding a future influenza pandemic.
On this latter and more-challenging front, there’s some encouraging news that was published recently in the journal Science [1]. An NIH-funded team has developed a unique “universal flu vaccine” that, with one seasonal shot, that has the potential to build immune protection against any of the 20 known subtypes of influenza virus and protect against future outbreaks.
While this experimental flu vaccine hasn’t yet been tested in people, the concept has shown great promise in advanced pre-clinical studies. Human clinical trials will hopefully start in the coming year. The researchers don’t expect that this universal flu vaccine will prevent influenza infection altogether. But, like COVID-19 vaccines, the new flu vaccine should help to reduce severe influenza illnesses and deaths when a person does get sick.
So, how does one develop a 20-in-1“multivalent” flu vaccine? It turns out that the key is the same messenger RNA (mRNA) technology that’s enabled two of the safe and effective vaccines against COVID-19, which have been so instrumental in fighting the pandemic. This includes the latest boosters from both Pfizer and Moderna, which now offer updated protection against currently circulating Omicron variants.
While this isn’t the first attempt to develop a universal flu vaccine, past attempts had primarily focused on a limited number of conserved antigens. An antigen is a protein or other substance that produces an immune response. Conserved antigens are those that tend to stay the same over time.
Because conserved antigens will look similar in many different influenza viruses, the hope was that vaccines targeting a small number of them would afford some broad influenza protection. But the focus on a strategy involving few antigens was driven largely by practical limitations. Using traditional methods to produce vaccines by growing flu viruses in eggs and isolating proteins, it simply isn’t feasible to include more than about four targets.
That’s where recent advances in mRNA technology come in. What makes mRNA so nifty for vaccines is that all you need to know is the letters, or sequence, that encodes the genetic material of a virus, including the sequences that get translated into proteins.
A research team led by Scott Hensley, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, recognized that the ease of designing and manufacturing mRNA vaccines opened the door to an alternate approach to developing a universal flu vaccine. Rather than limiting themselves to a few antigens, the researchers could make an all-in-one influenza vaccine, encoding antigens from every known influenza virus subtype.
Influenza vaccines generally target portions of a plentiful protein on the viral surface known as hemagglutinin (H). In earlier work, Hensley’s team, in collaboration with Perelman’s mRNA vaccine pioneer Drew Weissman, showed they could use mRNA technology to produce vaccines with H antigens from single influenza viruses [2, 3]. To protect the fragile mRNA molecules that encode a selected H antigen, researchers deliver them to cells inside well-tolerated microscopic lipid shells, or nanoparticles. The same is true of mRNA COVID-19 vaccines. In their earlier studies, the researchers found that when an mRNA vaccine aimed at one flu virus subtype was given to mice and ferrets in the lab, their cells made the encoded H antigen, eliciting protective antibodies.
In this latest study, they threw antigens from all 20 known flu viruses into the mix. This included H antigens from 18 known types of influenza A and two lineages of influenza B. The goal was to develop a vaccine that could teach the immune system to recognize and respond to any of them.
More study is needed, of course, but early indications are encouraging. The vaccine generated strong and broad antibody responses in animals. Importantly, it worked both in animals with no previous immunity to the flu and in those previously infected with flu viruses. That came as good news because past infections and resulting antibodies sometimes can interfere with the development of new antibodies against related viral subtypes.
In more good news, the researchers found that vaccinated mice and ferrets were protected against severe illness when later challenged with flu viruses. Those viruses included some that were closely matched to antigens in the vaccine, along with some that weren’t.
The findings offer proof-of-principle that mRNA vaccines containing a wide range of antigens can offer broad protection against influenza and likely other viruses as well, including the coronavirus strains responsible for COVID-19. The researchers report that they’re moving toward clinical trials in people, with the goal of beginning an early phase 1 trial in the coming year. The hope is that these developments—driven in part by technological advances and lessons learned over the course of the COVID-19 pandemic—will help to mitigate or perhaps even prevent future pandemics.
References:
[1] A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Arevalo CP, Bolton MJ, Le Sage V, Ye N, Furey C, Muramatsu H, Alameh MG, Pardi N, Drapeau EM, Parkhouse K, Garretson T, Morris JS, Moncla LH, Tam YK, Fan SHY, Lakdawala SS, Weissman D, Hensley SE. Science. 2022 Nov 25;378(6622):899-904.
[2] Nucleoside-modified mRNA vaccination partially overcomes maternal antibody inhibition of de novo immune responses in mice. Willis E, Pardi N, Parkhouse K, Mui BL, Tam YK, Weissman D, Hensley SE. Sci Transl Med. 2020 Jan 8;12(525):eaav5701.
[3] Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Pardi N, Parkhouse K, Kirkpatrick E, McMahon M, Zost SJ, Mui BL, Tam YK, Karikó K, Barbosa CJ, Madden TD, Hope MJ, Krammer F, Hensley SE, Weissman D. Nat Commun. 2018 Aug 22;9(1):3361.
Links:
Understanding Flu Viruses (Centers for Disease Control and Prevention, Atlanta)
COVID Research (NIH)
Decades in the Making: mRNA COVID-19 Vaccines (NIH)
Video: mRNA Flu Vaccines: Preventing the Next Pandemic (Penn Medicine, Philadelphia)
Scott Hensley (Perelman School of Medicine at the University of Pennsylvania, Philadelphia)
Weissman Lab (Perelman School of Medicine)
Video: The Story Behind mRNA COVID Vaccines: Katalin Karikó and Drew Weissman (Penn Medicine, Philadelphia)
NIH Support: National Institute for Allergy and Infectious Diseases
Posted In: News
Tags: coronavirus, COVID-19, COVID-19 booster, ferrets, flu, flu shot, flu virus, H antigens, hemagglutinin, human leukocyte antigen, influenza, influenza pandemic, influenza type A, influenza type B, influenza vaccine, influenza virus, mice, mRNA, mRNA vaccine, mRNA-lipid, nanoparticles, pandemic, universal flu vaccine, vaccine
Will Warm Weather Slow Spread of Novel Coronavirus?
Posted on June 2nd, 2020 by Dr. Francis Collins

Credit: Modified from iStock/energyy
With the start of summer coming soon, many are hopeful that the warmer weather will slow the spread of SARS-CoV-2, the novel coronavirus that causes COVID-19. There have been hints from lab experiments that increased temperature and humidity may reduce the viability of SARS-CoV-2. Meanwhile, other coronaviruses that cause less severe diseases, such as the common cold, do spread more slowly among people during the summer.
We’ll obviously have to wait a few months to get the data. But for now, many researchers have their doubts that the COVID-19 pandemic will enter a needed summertime lull. Among them are some experts on infectious disease transmission and climate modeling, who ran a series of sophisticated computer simulations of how the virus will likely spread over the coming months [1]. This research team found that humans’ current lack of immunity to SARS-CoV-2—not the weather—will likely be a primary factor driving the continued, rapid spread of the novel coronavirus this summer and into the fall.
These sobering predictions, published recently in the journal Science, come from studies led by Rachel Baker and Bryan Grenfell at Princeton Environmental Institute, Princeton, NJ. The Grenfell lab has long studied the dynamics of infectious illnesses, including seasonal influenza and respiratory syncytial virus (RSV). Last year, they published one of the first studies to look at how our warming climate might influence those dynamics in the coming years [2].
Those earlier studies focused on well-known human infectious diseases. Less clear is how seasonal variations in the weather might modulate the spread of a new virus that the vast majority of people and their immune systems have yet to encounter.
In the new study, the researchers developed a mathematical model to simulate how seasonal changes in temperature might influence the trajectory of COVID-19 in cities around the world. Of course, because the virus emerged on the scene only recently, we don’t know very much about how it will respond to warming conditions. So, the researchers ran three different scenarios based on what’s known about the role of climate in the spread of other viruses, including two coronaviruses, called OC43 and HKU1, that are known to cause common colds in people.
In all three scenarios, their models showed that climate only would become an important seasonal factor in controlling COVID-19 once a large proportion of people within a given community are immune or resistant to infection. In fact, the team found that, even if one assumes that SARS-CoV-2 is as sensitive to climate as other seasonal viruses, summer heat still would not be enough of a mitigator right now to slow its initial, rapid spread through the human population. That’s also clear from the rapid spread of COVID-19 that’s currently occurring in Brazil, Ecuador, and some other tropical nations.
Over the longer term, as more people develop immunity, the researchers suggest that COVID-19 may likely fall into a seasonal pattern similar to those seen with diseases caused by other coronaviruses. Long before then, NIH is working intensively with partners from all sectors to make sure that safe, effective treatments and vaccines will be available to help prevent the tragic, heavy loss of life that we’re seeing now.
Of course, climate is just one key factor to consider in evaluating the course of this disease. And, there is a glimmer of hope in one of the group’s models. The researchers incorporated the effects of control measures, such as physical distancing, with climate. It appears from this model that such measures, in combination with warm temperatures, actually might combine well to help slow the spread of this devastating virus. It’s a reminder that physical distancing will remain our best weapon into the summer to slow or prevent the spread of COVID-19. So, keep wearing those masks and staying 6 feet or more apart!
References:
[1] Susceptible supply limits the role of climate in the early SARS-CoV-2 pandemic. Baker RE, Yang W, Vecchi GA, Metcalf CJE, Grenfell BT. Science. 2020 May 18. [Online ahead of print.]
[2] Epidemic dynamics of respiratory syncytial virus in current and future climates. Baker RE, Mahmud AS, Wagner CE, Yang W, Pitzer VE, Viboud C, Vecchi GA, Metcalf CJE, Grenfell BT.Nat Commun. 2019 Dec 4;10(1):5512.
Links:
Coronavirus (COVID-19) (NIH)
Bryan Grenfell (Princeton University, Princeton, NJ)
Rachel Baker (Princeton University, Princeton, NJ)
Posted In: News
Tags: Brazil, climate, climate modeling, climate science, common cold, computer simulations, coronavirus, COVID-19, disease transmission, Ecuador, heat, HKU1 coronavirus, humidity, infectious disease, influenza, novel coronavirus, OC43 coronavirus, pandemic, physical distancing, respiratory syncytial virus, RSV, SARS-CoV-2, seasonal, social distancing, summer, summer camp, tropical
Can Smart Phone Apps Help Beat Pandemics?
Posted on April 9th, 2020 by Dr. Francis Collins

iStock/peterhowell
In recent weeks, most of us have spent a lot of time learning about coronavirus disease 2019 (COVID-19) and thinking about what’s needed to defeat this and future pandemic threats. When the time comes for people to come out of their home seclusion, how will we avoid a second wave of infections? One thing that’s crucial is developing better ways to trace the recent contacts of individuals who’ve tested positive for the disease-causing agent—in this case, a highly infectious novel coronavirus.
Traditional contact tracing involves a team of public health workers who talk to people via the phone or in face-to-face meetings. This time-consuming, methodical process is usually measured in days, and can even stretch to weeks in complex situations with multiple contacts. But researchers are now proposing to take advantage of digital technology to try to get contact tracing done much faster, perhaps in just a few hours.
Most smart phones are equipped with wireless Bluetooth technology that creates a log of all opt-in mobile apps operating nearby—including opt-in apps on the phones of nearby people. This has prompted a number of research teams to explore the idea of creating an app to notify individuals of exposure risk. Specifically, if a smart phone user tests positive today for COVID-19, everyone on their recent Bluetooth log would be alerted anonymously and advised to shelter at home. In fact, in a recent paper in the journal Science, a British research group has gone so far to suggest that such digital tracing may be valuable in the months ahead to improve our chances of keeping COVID-19 under control [1].
The British team, led by Luca Ferretti, Christophe Fraser, and David Bonsall, Oxford University, started their analyses using previously published data on COVID-19 outbreaks in China, Singapore, and aboard the Diamond Princess cruise ship. With a focus on prevention, the researchers compared the different routes of transmission, including from people with and without symptoms of the infection.
Based on that data, they concluded that traditional contact tracing was too slow to keep pace with the rapidly spreading COVID-19 outbreaks. During the three outbreaks studied, people infected with the novel coronavirus had a median incubation period of about five days before they showed any symptoms of COVID-19. Researchers estimated that anywhere from one-third to one-half of all transmissions came from asymptomatic people during this incubation period. Moreover, assuming that symptoms ultimately arose and an infected person was then tested and received a COVID-19 diagnosis, public health workers would need at least several more days to perform the contact tracing by traditional means. By then, they would have little chance of getting ahead of the outbreak by isolating the infected person’s contacts to slow its rate of transmission.
When they examined the situation in China, the researchers found that available data show a correlation between the roll-out of smart phone contact-tracing apps and the emergence of what appears to be sustained suppression of COVID-19 infection. Their analyses showed that the same held true in South Korea, where data collected through a smart phone app was used to recommend quarantine.
Despite its potential benefits in controlling or even averting pandemics, the British researchers acknowledged that digital tracing poses some major ethical, legal, and social issues. In China, people were required to install the digital tracing app on their phones if they wanted to venture outside their immediate neighborhoods. The app also displayed a color-coded warning system to enforce or relax restrictions on a person’s movements around a city or province. The Chinese app also relayed to a central database the information that it had gathered on phone users’ movements and COVID-19 status, raising serious concerns about data security and privacy of personal information.
In their new paper, the Oxford team, which included a bioethicist, makes the case for increased social dialogue about how best to employ digital tracing in ways the benefit human health. This is a far-reaching discussion with implications far beyond times of pandemic. Although the team analyzed digital tracing data for COVID-19, the algorithms that drive these apps could be adapted to track the spread of other common infectious diseases, such as seasonal influenza.
The study’s authors also raised another vital point. Even the most-sophisticated digital tracing app won’t be of much help if smart phone users don’t download it. Without widespread installation, the apps are unable to gather enough data to enable effective digital tracing. Indeed, the researchers estimate that about 60 percent of new COVID-19 cases in a community would need to be detected–and roughly the same percentage of contacts traced—to squelch the spread of the deadly virus.
Such numbers have app designers working hard to discover the right balance between protecting public health and ensuring personal rights. That includes NIH grantee Trevor Bedford, Fred Hutchinson Cancer Research Center, Seattle. He and his colleagues just launched NextTrace, a project that aims to build an opt-in app community for “digital participatory contact tracing” of COVID-19. Here at NIH, we have a team that is actively exploring the kind of technology that could achieve the benefits without unduly compromising personal privacy.
Bedford emphasizes that he and his colleagues aren’t trying to duplicate efforts already underway. Rather, they want to collaborate with others help to build a scientifically and ethically sound foundation for digital tracing aimed at improving the health of all humankind.
Reference:
[1] Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Ferretti L, Wymant C, Kendall M, Zhao L, Nurtay A, Abeler-Dörner L, Parker M, Bonsall D, Fraser C. Science. 2020 Mar 31. [Epub ahead of print]
Links:
Coronavirus (COVID-19) (NIH)
COVID-19, MERS & SARS (National Institute of Allergy and Infectious Diseases/NIH)
NextTrace (Fred Hutchinson Cancer Research Center, Seattle)
Bedford Lab (Fred Hutchinson Cancer Research Center)
NIH Support: National Institute of General Medical Sciences
Posted In: News
Tags: app, bioethics, Bluetooth, cell phone, China, contact tracing, coronavirus, COVID-19, crowdsourcing, data security, Diamond Princess, digital technologies, digital tracing, epidemic, epidemiology, infectious disease, influenza, mHealth, mobile health, NextTrace, novel coronavirus, opt-in app, pandemic, personal privacy, Singapore, smart phone, South Korea
How Measles Leave the Body Prone to Future Infections
Posted on November 12th, 2019 by Dr. Francis Collins

Credit: gettyimages/CHBD
As a kid who was home-schooled on a Virginia farm in the 1950s, I wasn’t around other kids very much, and so didn’t get exposed to measles. And there was no vaccine yet. Later on as a medical resident, I didn’t recognize that I wasn’t immune. So when I was hospitalized with a severe febrile illness at age 29, it took a while to figure out the diagnosis. Yes, it was measles. I have never been that sick before or since. I was lucky not to have long-term consequences, and now I’m learning that there may be even more to consider.
With the big push to get kids vaccinated, you’ve probably heard about some of the very serious complications of measles: hearing-threatening ear infections, bronchitis, laryngitis, and even life-threatening forms of pneumonia and encephalitis. But now comes word of yet another way in which the measles can be devastating—one that may also have long-term consequences for a person’s health.
In a new study in the journal Science, a research team, partly funded by NIH, found that the measles virus not only can make children deathly ill, it can cause their immune systems to forget how to ward off other common infections [1]. The virus does this by wiping out up to nearly three-quarters of the protective antibodies that a child’s body has formed in response to past microbial invaders and vaccinations. This immune “amnesia” can leave a child more vulnerable to re-contracting infections, such as influenza or respiratory syncytial virus (RSV), that they may have been protected against before they came down with measles.
The finding comes as yet another reason to feel immensely grateful that, thanks to our highly effective vaccination programs, most people born in the U.S. from the 1960s onward should never have to experience the measles.
There had been hints that the measles virus might somehow suppress a person’s immune system. Epidemiological evidence also had suggested that measles infections might lead to increased susceptibility to infection for years afterwards [2]. Scientists had even suspected this might be explained by a kind of immune amnesia. The trouble was that there wasn’t any direct proof that such a phenomenon actually existed.
In the new work, the researchers, led by Michael Mina, Tomasz Kula, and Stephen Elledge, Howard Hughes Medical Institute and Brigham and Women’s Hospital, Boston, took advantage of a tool developed a few years ago in the Elledge lab called VirScan [3]. VirScan detects antibodies in blood samples acquired as a result of a person’s past encounters with hundreds of viruses, bacteria, or vaccines, providing a comprehensive snapshot of acquired immunity at a particular moment in time.
To look for evidence of immune amnesia following the measles, the research team needed blood samples gathered from people both before and after infection. These types of samples are currently hard to come by in the U.S. thanks to the success of vaccines. By partnering with Rik de Swart, Erasmus University Medical Center, Rotterdam, Netherlands, they found the samples that they needed.
During a recent measles outbreak in the Netherlands, de Swart had gathered blood samples from children living in communities with low vaccination rates. Elledge’s group used VirScan with 77 unvaccinated kids to measure antibodies in samples collected before and about two months after their measles infections.
That included 34 children who had mild infections and 43 who had severe measles. The researchers also examined blood samples from five children who remained uninfected and 110 kids who hadn’t been exposed to the measles virus.
The VirScan data showed that the infected kids, not surprisingly, produced antibodies to the measles virus. But their other antibodies dropped and seemed to be disappearing. In fact, depending on the severity of measles infection, the kids showed on average a loss of around 40 percent of their antibody memory, with greater losses in children with severe cases of the measles. In at least one case, the loss reached a whopping 73 percent.
This all resonates with me. I do recall that after my bout with the measles, I seemed to be coming down with a lot of respiratory infections. I attributed that to the lifestyle of a medical resident—being around lots of sick patients and not getting much sleep. But maybe it was more than that.
The researchers suggest that the loss of immune memory may stem from the measles virus destroying some of the long-lived cells in bone marrow. These cells remember past infections and, based on that immunological memory, churn out needed antibodies to thwart reinvading viruses.
Interestingly, after a measles infection, the children’s immune systems still responded to new infections and could form new immune memories. But it appears the measles caused long term, possibly permanent, losses of a significant portion of previously acquired immunities. This loss of immune memory put the children at a distinct disadvantage should those old bugs circulate again.
It’s important to note that, unlike measles infection, the MMR (measles, mumps, rubella) vaccine does NOT compromise previously acquired immunity. So, these findings come as yet another reminder of the public value of measles vaccination.
Prior to 1963, when the measles vaccine was developed, 3 to 4 million Americans got the measles each year. As more people were vaccinated, the incidence of measles plummeted. By the year 2000, the disease was declared eliminated from the U.S.
Unfortunately, measles has made a come back, fueled by vaccine refusals. In October, the Centers for Disease Control and Prevention (CDC) reported an estimated 1,250 measles cases in the United States so far in 2019, surpassing the total number of cases reported annually in each of the past 25 years [4].
Around the world, measles continues to infect 7 million people each year, leading to an estimated 120,000 deaths. Based on the new findings, Elledge’s team now suspects the actual toll of the measles may be five times greater, due to the effects of immune amnesia.
The good news is those numbers can be reduced if more people get the vaccine, which has been shown repeatedly in many large and rigorous studies to be safe and effective. The CDC recommends that children should receive their first dose by 12 to 15 months of age and a second dose between the ages of 4 and 6. Older people who’ve been vaccinated or have had the measles previously should consider being re-vaccinated, especially if they live in places with low vaccination rates or will be traveling to countries where measles are endemic.
References:
[1] Measles virus infection diminishes preexisting antibodies that offer protection from other pathogens. Mina MJ, Kula T, Leng Y, Li M, de Vries RD, Knip M, Siljander H, Rewers M, Choy DF, Wilson MS, Larman HB, Nelson AN, Griffin DE, de Swart RL, Elledge SJ. et al. Science. 2019 Nov 1; 366 (6465): 599-606.
[2] Long-term measles-induced immunomodulation increases overall childhood infectious disease mortality. Mina MJ, Metcalf CJE, De Swart RL, Osterhaus ADME, Grenfell BT. Science. 2015 May 8; 348(6235).
[3] Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung’u T, Ruxrungtham K, Sanchez J, Brander C, Chung RT, O’Connor KC, Walker B, Larman HB, Elledge SJ. Science. 2015 Jun 5;348(6239):aaa0698.
[4] Measles cases and outbreaks. Centers for Disease Control and Prevention. Oct. 11, 2019.
Links:
Measles (MedlinePlus Medical Encyclopedia/National Library of Medicine/NIH)
Measles History (Centers for Disease Control and Prevention)
Vaccines (National Institute of Allergy and Infectious Diseases/NIAID)
Vaccines Protect Your Community (Vaccines.gov)
Elledge Lab (Harvard Medical School, Boston)
NIH Support: National Institute of Allergy and Infectious Diseases; National Institute of Diabetes and Digestive and Kidney Diseases
Posted In: News
Tags: antibodies, bone marrow, child health, childhood infectious diseases, immune amnesia, immunity, infection, infectious disease, influenza, measles, MMR vaccine, Netherlands, respiratory syncytial virus, RSV, vaccination, vaccine, VirScan, virus
These Oddball Cells May Explain How Influenza Leads to Asthma
Posted on May 16th, 2019 by Dr. Francis Collins
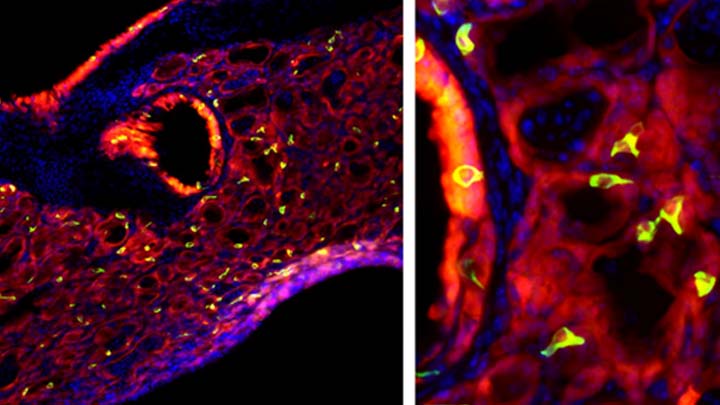
Credit: Andrew Vaughan, University of Pennsylvania, Philadelphia
Most people who get the flu bounce right back in a week or two. But, for others, the respiratory infection is the beginning of lasting asthma-like symptoms. Though I had a flu shot, I had a pretty bad respiratory illness last fall, and since that time I’ve had exercise-induced asthma that has occasionally required an inhaler for treatment. What’s going on? An NIH-funded team now has evidence from mouse studies that such long-term consequences stem in part from a surprising source: previously unknown lung cells closely resembling those found in taste buds.
The image above shows the lungs of a mouse after a severe case of H1N1 influenza infection, a common type of seasonal flu. Notice the oddball cells (green) known as solitary chemosensory cells (SCCs). Those little-known cells display the very same chemical-sensing surface proteins found on the tongue, where they allow us to sense bitterness. What makes these images so interesting is, prior to infection, the healthy mouse lungs had no SCCs.
SCCs, sometimes called “tuft cells” or “brush cells” or “type II taste receptor cells”, were first described in the 1920s when a scientist noticed unusual looking cells in the intestinal lining [1] Over the years, such cells turned up in the epithelial linings of many parts of the body, including the pancreas, gallbladder, and nasal passages. Only much more recently did scientists realize that those cells were all essentially the same cell type. Owing to their sensory abilities, these epithelial cells act as a kind of lookout for signs of infection or injury.
This latest work on SCCs, published recently in the American Journal of Physiology–Lung Cellular and Molecular Physiology, adds to this understanding. It comes from a research team led by Andrew Vaughan, University of Pennsylvania School of Veterinary Medicine, Philadelphia [2].
As a post-doc, Vaughan and colleagues had discovered a new class of cells, called lineage-negative epithelial progenitors, that are involved in abnormal remodeling and regrowth of lung tissue after a serious respiratory infection [3]. Upon closer inspection, they noticed that the remodeling of lung tissue post-infection often was accompanied by sustained inflammation. What they didn’t know was why.
The team, including Noam Cohen of Penn’s Perelman School of Medicine and De’Broski Herbert, also of Penn Vet, noticed signs of an inflammatory immune response several weeks after an influenza infection. Such a response in other parts of the body is often associated with allergies and asthma. All were known to involve SCCs, and this begged the question: were SCCs also present in the lungs?
Further work showed not only were SCCs present in the lungs post-infection, they were interspersed across the tissue lining. When the researchers exposed the animals’ lungs to bitter compounds, the activated SCCs multiplied and triggered acute inflammation.
Vaughan’s team also found out something pretty cool. The SCCs arise from the very same lineage of epithelial progenitor cells that Vaughan had discovered as a post-doc. These progenitor cells produce cells involved in remodeling and repair of lung tissue after a serious lung infection.
Of course, mice aren’t people. The researchers now plan to look in human lung samples to confirm the presence of these cells following respiratory infections.
If confirmed, the new findings might help to explain why kids who acquire severe respiratory infections early in life are at greater risk of developing asthma. They suggest that treatments designed to control these SCCs might help to treat or perhaps even prevent lifelong respiratory problems. The hope is that ultimately it will help to keep more people breathing easier after a severe bout with the flu.
References:
[1] Closing in on a century-old mystery, scientists are figuring out what the body’s ‘tuft cells’ do. Leslie M. Science. 2019 Mar 28.
[2] Development of solitary chemosensory cells in the distal lung after severe influenza injury. Rane CK, Jackson SR, Pastore CF, Zhao G, Weiner AI, Patel NN, Herbert DR, Cohen NA, Vaughan AE. Am J Physiol Lung Cell Mol Physiol. 2019 Mar 25.
[3] Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, Matthay MA, Rock JR, Chapman HA. Nature. 2015 Jan 29;517(7536):621-625.
Links:
Asthma (National Heart, Lung, and Blood Institute/NIH)
Influenza (National Institute of Allergy and Infectious Diseases/NIH)
Vaughan Lab (University of Pennsylvania, Philadelphia)
Cohen Lab (University of Pennsylvania, Philadelphia)
Herbert Lab (University of Pennsylvania, Philadelphia)
NIH Support: National Heart, Lung, and Blood Institute; National Institute on Deafness and Other Communication Disorders
Posted In: News
Tags: asthma, brush cells, cell biology, flu, H1N1, inflammation, influenza, lungs, nasal passage, nose, progenitor cells, respiratory infections, SCC, seasonal flu, solitary chemosensory cells, taste, tuft cells
Looking to Llamas for New Ways to Fight the Flu
Posted on November 13th, 2018 by Dr. Francis Collins
 Researchers are making tremendous strides toward developing better ways to reduce our risk of getting the flu. And one of the latest ideas for foiling the flu—a “gene mist” that could be sprayed into the nose—comes from a most surprising source: llamas.
Researchers are making tremendous strides toward developing better ways to reduce our risk of getting the flu. And one of the latest ideas for foiling the flu—a “gene mist” that could be sprayed into the nose—comes from a most surprising source: llamas.
Like humans and many other creatures, these fuzzy South American relatives of the camel produce immune molecules, called antibodies, in their blood when exposed to viruses and other foreign substances. Researchers speculated that because the llama’s antibodies are so much smaller than human antibodies, they might be easier to use therapeutically in fending off a wide range of flu viruses. This idea is now being leveraged to design a new type of gene therapy that may someday provide humans with broader protection against the flu [1].
Posted In: News
Tags: adeno-associated virus, antibody, antibody treatment, bioengineering, bird flu, cryo-EM, flu, flu pandemic, flu shot, gene mist, gene therapy, hemagglutinin, imaging, immunity, influenza, influenza type A, influenza type B, llama, nanobody, virus, x-ray crystallography
Seven More Awesome Technologies Made Possible by Your Tax Dollars
Posted on May 31st, 2018 by Dr. Francis Collins
We live in a world energized by technological advances, from that new app on your smartphone to drones and self-driving cars. As you can see from this video, NIH-supported researchers are also major contributors, developing a wide range of amazing biomedical technologies that offer tremendous potential to improve our health.
Produced by the NIH’s National Institute of Biomedical Imaging and Bioengineering (NIBIB), this video starts by showcasing some cool fluorescent markers that are custom-designed to light up specific cells in the body. This technology is already helping surgeons see and remove tumor cells with greater precision in people with head and neck cancer [1]. Further down the road, it might also be used to light up nerves, which can be very difficult to see—and spare—during operations for cancer and other conditions.
Other great things to come include:
- A wearable tattoo that detects alcohol levels in perspiration and wirelessly transmits the information to a smartphone.
- Flexible coils that produce high quality images during magnetic resonance imaging (MRI) [2-3]. In the future, these individualized, screen-printed coils may improve the comfort and decrease the scan times of people undergoing MRI, especially infants and small children.
- A time-release capsule filled with a star-shaped polymer containing the anti-malarial drug ivermectin. The capsule slowly dissolves in the stomach over two weeks, with the goal of reducing the need for daily doses of ivermectin to prevent malaria infections in at-risk people [4].
- A new radiotracer to detect prostate cancer that has spread to other parts of the body. Early clinical trial results show the radiotracer, made up of carrier molecules bonded tightly to a radioactive atom, appears to be safe and effective [5].
- A new supercooling technique that promises to extend the time that organs donated for transplantation can remain viable outside the body [6-7]. For example, current technology can preserve donated livers outside the body for just 24 hours. In animal studies, this new technique quadruples that storage time to up to four days.
- A wearable skin patch with dissolvable microneedles capable of effectively delivering an influenza vaccine. This painless technology, which has produced promising early results in humans, may offer a simple, affordable alternative to needle-and-syringe immunization [8].
If you like what you see here, be sure to check out this previous NIH video that shows six more awesome biomedical technologies that your tax dollars are helping to create. So, let me extend a big thanks to you from those of us at NIH—and from all Americans who care about the future of their health—for your strong, continued support!
References:
[1] Image-guided surgery in cancer: A strategy to reduce incidence of positive surgical margins. Wiley Interdiscip Rev Syst Biol Med. 2018 Feb 23.
[2] Screen-printed flexible MRI receive coils. Corea JR, Flynn AM, Lechêne B, Scott G, Reed GD, Shin PJ, Lustig M, Arias AC. Nat Commun. 2016 Mar 10;7:10839.
[3] Printed Receive Coils with High Acoustic Transparency for Magnetic Resonance Guided Focused Ultrasound. Corea J, Ye P, Seo D, Butts-Pauly K, Arias AC, Lustig M. Sci Rep. 2018 Feb 21;8(1):3392.
[4] Oral, ultra-long-lasting drug delivery: Application toward malaria elimination goals. Bellinger AM, Jafari M1, Grant TM, Zhang S, Slater HC, Wenger EA, Mo S, Lee YL, Mazdiyasni H, Kogan L, Barman R, Cleveland C, Booth L, Bensel T, Minahan D, Hurowitz HM, Tai T, Daily J, Nikolic B, Wood L, Eckhoff PA, Langer R, Traverso G. Sci Transl Med. 2016 Nov 16;8(365):365ra157.
[5] Clinical Translation of a Dual Integrin avb3– and Gastrin-Releasing Peptide Receptor–Targeting PET Radiotracer, 68Ga-BBN-RGD. Zhang J, Niu G, Lang L, Li F, Fan X, Yan X, Yao S, Yan W, Huo L, Chen L, Li Z, Zhu Z, Chen X. J Nucl Med. 2017 Feb;58(2):228-234.
[6] Supercooling enables long-term transplantation survival following 4 days of liver preservation. Berendsen TA, Bruinsma BG, Puts CF, Saeidi N, Usta OB, Uygun BE, Izamis ML, Toner M, Yarmush ML, Uygun K. Nat Med. 2014 Jul;20(7):790-793.
[7] The promise of organ and tissue preservation to transform medicine. Giwa S, Lewis JK, Alvarez L, Langer R, Roth AE, et a. Nat Biotechnol. 2017 Jun 7;35(6):530-542.
[8] The safety, immunogenicity, and acceptability of inactivated influenza vaccine delivered by microneedle patch (TIV-MNP 2015): a randomised, partly blinded, placebo-controlled, phase 1 trial. Rouphael NG, Paine M, Mosley R, Henry S, McAllister DV, Kalluri H, Pewin W, Frew PM, Yu T, Thornburg NJ, Kabbani S, Lai L, Vassilieva EV, Skountzou I, Compans RW, Mulligan MJ, Prausnitz MR; TIV-MNP 2015 Study Group.
Links:
National Institute of Biomedical Imaging and Bioengineering (NIH)
Center for Wearable Sensors (University of California, San Diego)
Hyperpolarized MRI Technology Resource Center (University of California, San Francisco)
Center for Engineering in Medicine (Massachusetts General Hospital, Boston)
Center for Drug Design, Development and Delivery (Georgia Tech University, Atlanta)
NIH Support: National Institute of Biomedical Imaging and Bioengineering; National Institute of Diabetes and Digestive and Kidney Diseases; National Institute of Allergy and Infectious Diseases
Posted In: Cool Videos
Tags: antimalarial drugs, bioengineering, cancer, cell phone, Cool Videos, drug delivery, head and neck cancer, image-guided surgery, imaging, influenza, influenza vaccine, ivermectin, liver transplant, magnetic resonance imaging, malaria, MRI, NIBIB, organ transplant, prostate cancer, radiotracer, screen-printed coils, smartphone, supercooling, technology, transplants, video
Creative Minds: Does Human Immunity Change with the Seasons?
Posted on March 9th, 2017 by Dr. Francis Collins
Micaela Martinez
It’s an inescapable conclusion from the book of Ecclesiastes that’s become part of popular culture thanks to folk legends Pete Seeger and The Byrds: “To everything (turn, turn, turn), there is a season.” That’s certainly true of viral outbreaks, from the flu-causing influenza virus peaking each year in the winter to polio outbreaks often rising in the summer. What fascinates Micaela Martinez is, while those seasonal patterns of infection have been recognized for decades, nobody really knows why they occur.
Martinez, an infectious disease ecologist at Princeton University, Princeton, NJ, thinks colder weather conditions and the tendency for humans to stay together indoors in winter surely play a role. But she also thinks an important part of the answer might be found in a place most hadn’t thought to look: seasonal changes in the human immune system. Martinez recently received an NIH Director’s 2016 Early Independence Award to explore fluctuations in the body’s biological rhythms over the course of the year and their potential influence on our health.
Tags: biological rhythms, Bridges to the Baccalaureate Program, chickenpox, circadian rhythms, cytomegalovirus, flu, herpes virus, immune system, immunity, immunobiology, infectious disease, infectious disease ecology, influenza, NIH Director’s 2016 Early Independence Award, seasonal flu, shingles, sleep, vaccine, varicella-zoster virus
Snapshots of Life: Virus Hunting with Carbon Nanotubes
Posted on December 1st, 2016 by Dr. Francis Collins

Credit: Penn State University
The purple pods that you see in this scanning electron micrograph are the H5N2 avian flu virus, a costly threat to the poultry and egg industry and, in very rare instances, a health risk for humans. However, these particular pods are unlikely to infect anything because they are trapped in a gray mesh of carbon nanotubes. Made by linking carbon atoms into a cylindrical pattern, such nanotubes are about 10,000 times smaller than width of a human hair.
The nanotubes above have been carefully aligned on a special type of silicon chip called a carbon-nanotube size-tunable-enrichment-microdevice (CNT-STEM). As described recently in Science Advances, this ultrasensitive device is designed to capture viruses rapidly based on their size, not their molecular characteristics [1]. This unique feature enables researchers to detect completely unknown viruses, even when they are present in extremely low numbers. In proof-of-principle studies, CNT-STEM made it possible to collect and detect viruses in a sample at concentrations 100 times lower than with other methods, suggesting the device and its new approach will be helpful in the ongoing hunt for new and emerging viruses, including those that infect people.
Tags: avian influenza, bird flu, carbon nanotubes, chemistry, CNT-STEM, diagnostics, ducks, genomics, H11N9 avian flu virus, H11N9 avian influenza, H5N2 avian flu virus, H5N2 avian influenza, infectious disease, influenza, materials science, microdevice, nanoengineering, nanotechnology, nanotube, NIH Director's New Innovator Award, physics, poultry, silicon chip microdevice, turkey, virology, virus
