lung cancer – NIH Director's Blog (original) (raw)
New Approach to ‘Liquid Biopsy’ Relies on Repetitive RNA in the Bloodstream
Posted on September 12th, 2023 by Lawrence Tabak, D.D.S., Ph.D.
Researchers have identified segments of noncoding RNA circulating in the blood that are early signs of cancer. Credit: Modified from Adobe Stock/ Andrey Popov; Donny Bliss, NIH
It’s always best to diagnose cancer at an early stage when treatment is most likely to succeed. Unfortunately, far too many cancers are still detected only after cancer cells have escaped from a primary tumor and spread to distant parts of the body. This explains why there’s been so much effort in recent years to develop liquid biopsies, which are tests that can pick up on circulating cancer cells or molecular signs of cancer in blood or other bodily fluids and reliably trace them back to the organ in which a potentially life-threatening tumor is growing.
Earlier methods to develop liquid biopsies for detecting cancers often have relied on the presence of cancer-related proteins and/or DNA in the bloodstream. Now, an NIH-supported research team has encouraging evidence to suggest that this general approach to detecting cancers—including aggressive pancreatic cancers—may work even better by taking advantage of signals from a lesser-known form of genetic material called noncoding RNA.
The findings reported in Nature Biomedical Engineering suggest that the new liquid biopsy approach may aid in the diagnosis of many forms of cancer [1]. The studies show that the sensitivity of the tests varies—a highly sensitive test is one that rarely misses cases of disease. However, they already have evidence that millions of circulating RNA molecules may hold promise for detecting cancers of the liver, esophagus, colon, stomach, and lung.
How does it work? The human genome contains about 3 billion paired DNA letters. Most of those letters are transcribed, or copied, into single-stranded RNA molecules. While RNA is best known for encoding proteins that do the work of the cell, most RNA never gets translated into proteins at all. This noncoding RNA includes repetitive RNA that can be transcribed from millions of repeat elements—patterns of the same few DNA letters occurring multiple times in the genome.
Common approaches to studying RNA don’t analyze repetitive RNA, so its usefulness as a diagnostic tool has been unclear—until recently. Last year, the lab of Daniel Kim at the University of California, Santa Cruz reported [2] that a key genetic mutation that occurs early on in some cancers causes repetitive RNA molecules to be secreted in large quantities from cancer cells, even at the earliest stages of cancer. Non-cancerous cells, by comparison, release much less repetitive RNA.
The findings suggested that liquid biopsy tests that look for this repetitive, noncoding RNA might offer a powerful new way to detect cancers sooner, according to the authors. But first they needed a method capable of measuring it. Due to its oftentimes uncertain functions, the researchers have referred to repetitive, noncoding RNA as “dark matter.”
Using a liquid biopsy platform they developed called COMPLETE-seq, Kim’s team trained computers to detect cancers by looking for patterns in RNA data. The platform enables sequencing and analysis of all protein coding and noncoding RNAs—including any RNA from more than 5 million repeat elements—present in a blood sample. They found that their classifiers worked better when repetitive RNAs were included. The findings lend support to the idea that repetitive, noncoding RNA in the bloodstream is a rich source of information for detecting cancers, which has previously been overlooked.
In a study comparing blood samples from healthy people to those with pancreatic cancer, the COMPLETE-seq technology showed that nearly all people in the study with pancreatic cancer had more repetitive, noncoding RNA in their blood samples compared to healthy people, according to the researchers. They used the COMPLETE-seq test on blood samples from people with other types of cancer as well. For example, their test accurately detected 91% of colorectal cancer samples and 93% of lung cancer samples.
They now plan to look at many more cancer types with samples from additional patients representing a broad range of cancer stages. The goal is to develop a single RNA liquid biopsy test that could detect multiple forms of cancer with a high degree of accuracy and specificity. They note that such a test might also be used to guide treatment decisions and more readily detect a cancer’s recurrence. The hope is that one day a comprehensive liquid biopsy test including coding and noncoding RNA will catch many more cancers sooner, when treatment can be most successful.
References:
[1] RE Reggiardo et al. Profiling of repetitive RNA sequences in the blood plasma of patients with cancer. Nature Biomedical Engineering DOI: 10.1038/s41551-023-01081-7 (2023).
[2] RE Reggiardo et al. Mutant KRAS regulates transposable element RNA and innate immunity via KRAB zinc-finger genes. Cell Reports DOI: 10.1016/j.celrep.2022.111104 (2022).
Links:
Daniel Kim Lab (UC Santa Cruz)
Cancer Screening Overview (National Cancer Institute/NIH)
Early Detection (National Cancer Institute/NIH)
NIH Support: National Cancer Institute, National Heart, Lung, and Blood Institute, National Institute of Diabetes and Digestive and Kidney Diseases
Posted In: Health, News, Science, technology
Tags: cancer, colorectal cancer, Daniel Kim, diagnosing cancer, liquid biopsy, lung cancer, noncoding RNA, pancreatic cancer, repetitive rna, RNA
Teaching the Immune System to Attack Cancer with Greater Precision
Posted on November 12th, 2021 by Dr. Francis Collins

Credit: PhotobyTawat/Shutterstock/Tom Deerink, National Institute of General Medical Sciences, NIH
To protect humans from COVID-19, the Pfizer and Moderna mRNA vaccines program human cells to translate the injected synthetic messenger RNA into the coronavirus spike protein, which then primes the immune system to arm itself against future appearances of that protein. It turns out that the immune system can also be trained to spot and attack distinctive proteins on cancer cells, killing them and leaving healthy cells potentially untouched.
While these precision cancer vaccines remain experimental, researchers continue to make basic discoveries that move the field forward. That includes a recent NIH-funded study in mice that helps to refine the selection of protein targets on tumors as a way to boost the immune response [1]. To enable this boost, the researchers first had to discover a possible solution to a longstanding challenge in developing precision cancer vaccines: T cell exhaustion.
The term refers to the immune system’s complement of T cells and their capacity to learn to recognize foreign proteins, also known as neoantigens, and attack them on cancer cells to shrink tumors. But these responding T cells can exhaust themselves attacking tumors, limiting the immune response and making its benefits short-lived.
In this latest study, published in the journal Cell, Tyler Jacks and Megan Burger, Massachusetts Institute of Technology, Cambridge, help to explain this phenomenon of T cell exhaustion. The researchers found in mice with lung tumors that the immune system initially responds as it should. It produces lots of T cells that target many different cancer-specific proteins.
Yet there’s a problem: various subsets of T cells get in each other’s way. They compete until, eventually, one of those subsets becomes the dominant T cell type. Even when those dominant T cells grow exhausted, they still remain in the tumor and keep out other T cells, which might otherwise attack different neoantigens in the cancer.
Building on this basic discovery, the researchers came up with a strategy for developing cancer vaccines that can “awaken” T cells and reinvigorate the body’s natural cancer-fighting abilities. The strategy might seem counterintuitive. The researchers vaccinated mice with neoantigens that provide a weak but encouraging signal to the immune cells responsible for presenting the distinctive cancer protein target, or antigen, to T cells. It’s those T cells that tend to get suppressed in competition with other T cells.
When the researchers vaccinated the mice with one of those neoantigens, the otherwise suppressed T cells grew in numbers and better targeted the tumor. What’s more, the tumors shrank by more than 25 percent on average.
Research on this new strategy remains in its early stages. The researchers hope to learn if this approach to cancer vaccines might work even better when used in combination with immunotherapy drugs, which unleash the immune system against cancer in other ways.
It’s also possible that the recent and revolutionary success of mRNA vaccines for preventing COVID-19 actually could help. An important advantage of mRNA is that it’s easy for researchers to synthesize once they know the specific nucleic acid sequence of a protein target, and they can even combine different mRNA sequences to make a multiplex vaccine that primes the immune system to recognize multiple neoantigens. Now that we’ve seen how well mRNA vaccines work to prompt a desired immune response against COVID-19, this same technology can be used to speed the development and testing of future vaccines, including those designed precisely to fight cancer.
Reference:
[1] Antigen dominance hierarchies shape TCF1+ progenitor CD8 T cell phenotypes in tumors. Burger ML, Cruz AM, Crossland GE, Gaglia G, Ritch CC, Blatt SE, Bhutkar A, Canner D, Kienka T, Tavana SZ, Barandiaran AL, Garmilla A, Schenkel JM, Hillman M, de Los Rios Kobara I, Li A, Jaeger AM, Hwang WL, Westcott PMK, Manos MP, Holovatska MM, Hodi FS, Regev A, Santagata S, Jacks T. Cell. 2021 Sep 16;184(19):4996-5014.e26.
Links:
Cancer Treatment Vaccines (National Cancer Institute/NIH)
The Jacks Lab (Massachusetts Institute of Technology, Cambridge)
NIH Support: National Cancer Institute; National Heart, Lung, and Blood Institute
Posted In: News
Tags: cancer, cancer immunotherapy, cancer treatment vaccines, cancer vaccine, immune system, immunology, lung cancer, mRNA, mRNA vaccines, neoantigen, precision cancer vaccine, precision medicine, T cell exhaustion, T cells
Tackling Cancer Metastasis with Engineered Blood Platelets
Posted on August 27th, 2020 by Dr. Francis Collins

Tara Deans
Credit: Dan Hixson/University of Utah College of Engineering, Salt Lake City
When cancer cells spread to new parts of the body in a process called metastasis, they often get there by traveling through the bloodstream. To avoid alerting the immune system and possibly triggering their demise, cancer cells coax circulating blood platelets to glom onto their surfaces and mask them from detection. This deceptive arrangement has raised a tantalizing possibility: What if blood platelets could be programmed to recognize and take out those metastasizing cancer cells?
Tara Deans, University of Utah, Salt Lake City, was recently awarded a 2019 NIH Director’s New Innovator Award to do exactly that. It’s an exciting opportunity for a researcher who stumbled onto this innovative strategy quite by accident.
Deans is a bioengineer and expert in designing synthetic gene circuits. These circuits consist of small collections of genetic “parts” that can be assembled and integrated to program cells to behave differently than their natural counterparts [1]. In her initial work, Deans got these specialized gene circuits to prompt blood-forming stem cells to mass-produce platelets in the lab.
But blood platelets are unusual cells. They’re packed with many proteins that help to repair small nicks in blood vessels and stop the bleeding when we’re injured. Blood platelets do so even though they lack a nucleus and DNA to encode and make any of the proteins. Their protein cargo is pre-packaged and comes strictly from the bone marrow cells, called megakaryocytes, that produce them.
Deans realized that engineering platelets might pose a rare opportunity. She could wire the needed circuitry into the blood-forming stem cells and engineer them to make any desired therapeutic proteins, which are then loaded into the blood platelets for their 8- to 10-day lifespan. She started out producing blood platelets that could safely carry functional replacement enzymes in people with certain rare metabolic disorders.
As this research progressed, Deans got some troubling personal news: A friend was diagnosed with a blood cancer. At the time, Deans didn’t know much about the diagnosis. But, in reading about her friend’s cancer, she learned how metastasizing tumor cells interact with platelets.
That’s when Deans had her “aha” moment: maybe the engineered platelets could also be put to work in preventing metastasizing tumor cells from spreading.
Now, with her New Innovator Award, Deans will pursue this novel approach by engineering platelets to carry potentially promising cancer-fighting proteins. In principle, they could be tailored to fight breast, lung, and various other cancer types. Ultimately, she hopes that platelets could be engineered to target and kill circulating cancer cells before they move into other tissues.
There’s plenty of research ahead to work out the details of targeting the circulating cancer cells and then testing them in animal models before this strategy could ever be attempted in people. But Deans is excited about the path forward, and thinks that platelets hold great promise to function as unique drug delivery devices. It has not escaped her notice that this approach could work not only for controlling the spread of cancer cells, but also in treating other medical conditions.
Reference:
[1] Genetic circuits to engineer tissues with alternative functions. Healy CP, Deans TL. J Biol Eng. 2019 May 3;13:39.
Links:
Metastatic Cancer (National Cancer Institute/NIH)
Deans Lab (University of Utah, Salt Lake City)
Deans Project Information (NIH RePORTER)
NIH Director’s New Innovator Award (Common Fund)
NIH Support: Common Fund; National Cancer Institute
Posted In: Creative Minds
Tags: 2019 NIH Director’s New Innovator Award, bioengineering, blood cancer, blood platelets, blood-forming stem cells, breast cancer, cancer, cancer metastasis, drug delivery, gene circuits, lung cancer, megakaryocytes, metabolic disorders, platelets, stem cells, synthetic biology, synthetic gene circuits
Working to Improve Immunotherapy for Lung Cancer
Posted on January 30th, 2020 by Dr. Francis Collins

Credit: Xiaodong Zhu, Fred Hutchinson Cancer Research Center, Seattle
For those who track cancer statistics, this year started off on a positive note with word that lung cancer deaths continue to decline in the United States [1]. While there’s plenty of credit to go around for that encouraging news—and continued reduction in smoking is a big factor—some of this progress likely can be ascribed to a type of immunotherapy, called PD-1 inhibitors. This revolutionary approach has dramatically changed the treatment landscape for the most common type of lung cancer, non-small cell lung cancer (NSCLC).
PD-1 inhibitors, which have only been available for about five years, prime one component of a patient’s own immune system, called T cells, to seek and destroy malignant cells in the lungs. Unfortunately, however, only about 20 percent of people with NSCLC respond to PD-1 inhibitors. So, many researchers, including the team of A. McGarry Houghton, Fred Hutchinson Cancer Research Center, Seattle, are working hard to extend the benefits of immunotherapy to more cancer patients.
The team’s latest paper, published in JCI Insight [2], reveals that one culprit behind a poor response to immunotherapy may be the immune system’s own first responders: neutrophils. Billions of neutrophils circulate throughout the body to track down abnormalities, such as harmful bacteria and malignant cells. They also contact other parts of the immune system, including T cells, if help is needed to eliminate the health threat.
In their study, the Houghton team, led by Julia Kargl, combined several lab techniques to take a rigorous, unbiased look at the immune cell profiles of tumor samples from dozens of NSCLC patients who received PD-1 inhibitors as a frontline treatment. The micrographs above show tumor samples from two of these patients.
In the image on the left, large swaths of T cells (light blue) have infiltrated the cancer cells (white specks). Interestingly, other immune cells, including neutrophils (magenta), are sparse.
In contrast, in the image on the right, T cells (light blue) are sparse. Instead, the tumor teems with other types of immune cells, including macrophages (red), two types of monocytes (yellow, green), and, most significantly, lots of neutrophils (magenta). These cells arise from myeloid progenitor cells in the bone marrow, while T cells arise from the marrow’s lymphoid progenitor cell.
Though the immune profiles of some tumor samples were tough to classify, the researchers found that most fit neatly into two subgroups: tumors showing active levels of T cell infiltration (like the image on the left) or those with large numbers of myeloid immune cells, especially neutrophils (like the image on the right). This dichotomy then served as a reliable predictor of treatment outcome. In the tumor samples with majority T cells, the PD-1 inhibitor worked to varying degrees. But in the tumor samples with predominantly neutrophil infiltration, the treatment failed.
Houghton’s team has previously found that many cancers, including NSCLC, actively recruit neutrophils, turning them into zombie-like helpers that falsely signal other immune cells, like T cells, to stay away. Based on this information, Houghton and colleagues used a mouse model of lung cancer to explore a possible way to increase the success rate of PD-1 immunotherapy.
In their mouse experiments, the researchers found that when PD-1 was combined with an existing drug that inhibits neutrophils, lung tumors infiltrated with neutrophils were converted into tumors infiltrated by T cells. The tumors treated with the combination treatment also expressed genes associated with an active immunotherapy response.
This year, January brought encouraging news about decreasing deaths from lung cancer. But with ongoing basic research, like this study, to tease out the mechanisms underlying the success and failure of immunotherapy, future months may bring even better news.
References:
[1] Cancer statistics, 2020. Siegel RL, Miller KD, Jemal A. CA Cancer J Clin. 2020 Jan;70(1):7-30.
[2] Neutrophil content predicts lymphocyte depletion and anti-PD1 treatment failure in NSCLC. Kargl J, Zhu X, Zhang H, Yang GHY, Friesen TJ, Shipley M, Maeda DY, Zebala JA, McKay-Fleisch J, Meredith G, Mashadi-Hossein A, Baik C, Pierce RH, Redman MW, Thompson JC, Albelda SM, Bolouri H, Houghton AM. JCI Insight. 2019 Dec 19;4(24).
[3] Neutrophils dominate the immune cell composition in non-small cell lung cancer. Kargl J, Busch SE, Yang GH, Kim KH, Hanke ML, Metz HE, Hubbard JJ, Lee SM, Madtes DK, McIntosh MW, Houghton AM. Nat Commun. 2017 Feb 1;8:14381.
Links:
Non-Small Cell Lung Cancer Treatment (PDQ®)–Patient Version (National Cancer Institute/NIH)
Spotlight on McGarry Houghton (Fred Hutchinson Cancer Research Center, Seattle)
Houghton Lab (Fred Hutchinson Cancer Research Center)
NIH Support: National Cancer Institute
Posted In: Snapshots of Life
Tags: cancer, cancer immunotherapy, immunology, lung cancer, lungs, lymphoid progenitor cells, macrophage, monocyte, myeloid progenitor cells, non-small cell lung cancer, NSCLC, PD-1 inhibitors, T cells
Study Suggests Repurposed Drugs Might Treat Aggressive Lung Cancer
Posted on November 19th, 2019 by Dr. Francis Collins

Caption: Small cell lung cancer cells (red) spreading via blood vessels (white) from the lung to the liver of a genetically-engineered mouse model.
Credit: Leanne Li, Koch Institute at MIT
Despite continued progress in treatment and prevention, lung cancer remains our nation’s leading cause of cancer death. In fact, more Americans die of lung cancer each year than of breast, colon, and prostate cancers combined [1,2]. While cigarette smoking is a major cause, lung cancer also occurs in non-smokers. I’m pleased to report discovery of what we hope will be a much-needed drug target for a highly aggressive, difficult-to-treat form of the disease, called small cell lung cancer (SCLC).
Using gene-editing technology to conduct a systematic, large-scale search for druggable vulnerabilities in certain types of cancer cells grown in lab dishes, NIH-funded researchers recently identified a metabolic pathway that appears to play a key role in SCLC. What makes this news even more encouraging is drugs that block this pathway already exist. That includes one in clinical testing for other types of cancer, and another that’s FDA-approved and has been safely used for more than 20 years to treat people with rheumatoid arthritis.
The new work comes from the lab of Tyler Jacks, Massachusetts Institute of Technology (MIT), Cambridge. The Jacks lab, which is dedicated to understanding the genetic events that lead to cancer, develops mouse models engineered to carry the same genetic mutations that turn up in human cancers.
In work described in Science Translational Medicine, the team, co-led by Leanne Li and Sheng Rong Ng, applied CRISPR gene-editing tools to cells grown from some of their mouse models. Aiming high in terms of scale, researchers used CRISPR to knock out systematically, one by one, each of about 5,000 genes in cells from the SCLC mouse model, as well in cells from mouse models of other types of lung and pancreatic cancers. They looked to see what gene knockouts would slow down or kill the cancer cells, because that would be a good indication that the protein products of these genes, or the pathways they mediated, would be potential drug targets.
Out of those thousands of genes, one rose to the top of the list. It encodes an enzyme called DHODH (dihydroorotate dehydrogenase). This enzyme plays an important role in synthesizing pyrimidine, which is a major building block in DNA and RNA. Cytosine and thymine, the C and T in the four-letter DNA code, are pyrimidines; so is uracil, the U in RNA that takes the place of T in DNA. Because cancer cells are constantly dividing, there is a continual need to synthesize new DNA and RNA molecules to support the production of new daughter cells. And that means, unlike healthy cells, cancer cells require a steady supply of pyrimidine.
It turns out that the SCLC cells have an unexpected weakness relative to other cancer cells: they don’t produce as much pyrimidine. As a result, the researchers found blocking DHODH left the cells short on pyrimidine, leading to reduced growth and survival of the cancer.
This was especially good news because DHODH-blocking drugs, including one called brequinar, have already been tested in clinical trials for other cancers. In fact, brequinar is now being explored as a potential treatment for acute myeloid leukemia.
Might brequinar also hold promise for treating SCLC? To explore further, the researchers looked again to their genetic mouse model of SCLC. Their studies showed that mice treated with brequinar lived about 40 days longer than control animals. That’s a significant survival benefit in this system.
Brequinar treatment appeared to work even better when combined with other approved cancer drugs in mice that had SCLC cells transplanted into them. Further study in mice carrying SCLC tumors derived from four human patients added to this evidence. Two of the four human tumors shrunk in mice treated with brequinar.
Of course, mice are not people. But the findings suggest that brequinar or another DHODH blocker might hold promise as a new way to treat SCLC. While more study is needed to understand even better how brequinar works and explore potentially promising drug combinations, the fact that this drug is already in human testing for another indication suggests that a clinical trial to explore its use for SCLC might happen more quickly.
More broadly, the new findings show the promise of gene-editing technology as a research tool for uncovering elusive cancer targets. Such hard-fought discoveries will help to advance precise approaches to the treatment of even the most aggressive cancer types. And that should come as encouraging news to all those who are hoping to find new answers for hard-to-treat cancers.
References:
[1] Cancer Stat Facts: Lung and Bronchus Cancer (National Cancer Institute/NIH)
[2] Key Statistics for Lung Cancer (American Cancer Society)
[3] Identification of DHODH as a therapeutic target in small cell lung cancer. Li L, Ng SR, Colón CI, Drapkin BJ, Hsu PP, Li Z, Nabel CS, Lewis CA, Romero R, Mercer KL, Bhutkar A, Phat S, Myers DT, Muzumdar MD, Westcott PMK, Beytagh MC, Farago AF, Vander Heiden MG, Dyson NJ, Jacks T. Sci Transl Med. 2019 Nov 6;11(517).
Links:
Small Cell Lung Cancer Treatment (NCI/NIH)
Video: Introduction to Genome Editing Using CRISPR Cas9 (NIH)
Tyler Jacks (Massachusetts Institute of Technology, Cambridge)
NIH Support: National Cancer Institute
Posted In: News
Tags: brequinar, cancer, CRISPR, DHODH, dihydroorotate dehydrogenase, drug target, gene editing, gene knockouts, lung cancer, mouse model, pyrimidine, SCLC, small cell lung cancer
Panel Finds Exercise May Lower Cancer Risk, Improve Outcomes
Posted on October 16th, 2019 by Dr. Francis Collins

Credit: gettyimages/vgajic
Exercise can work wonders for your health, including strengthening muscles and bones, and boosting metabolism, mood, and memory skills. Now comes word that staying active may also help to lower your odds of developing cancer.
After reviewing the scientific evidence, a panel of experts recently concluded that physical activity is associated with reduced risks for seven common types of cancer: colon, breast, kidney, endometrial, bladder, stomach, and esophageal adenocarcinoma. What’s more, the experts found that exercise—both before and after a cancer diagnosis—was linked to improved survival among people with breast, colorectal, or prostate cancers.
About a decade ago, the American College of Sports Medicine (ACSM) convened its first panel of experts to review the evidence on the role of exercise in cancer. At the time, there was limited evidence to suggest a connection between exercise and a reduced risk for breast, colon, and perhaps a few other cancer types. There also were some hints that exercise might help to improve survival among people with a diagnosis of cancer.
Today, the evidence linking exercise and cancer has grown considerably. That’s why the ACSM last year convened a group of 40 experts to perform a comprehensive review of the research literature and summarize the level of the evidence. The team, including Charles Matthews and Frank Perna with the NIH’s National Cancer Institute, reported its findings and associated guidelines and recommendations in three papers just published in Medicine & Science in Sports & Exercise and CA: A Cancer Journal for Clinicians [1,2,3].
Here are some additional highlights from the papers:
There’s moderate evidence to support an association between exercise and reduced risk for some other cancer types, including cancers of the lung and liver.
While the optimal amount of exercise needed to reduce cancer risk is still unclear, being physically active is clearly one of the most important steps in general that people of all ages and abilities can take.
Is sitting the new smoking? Reducing the amount of time spent sitting also may help to lower the risk of some cancers, including endometrial, colon, and lung cancers. However, there’s not enough evidence to draw clear conclusions yet.
Every cancer survivor should, within reason, “avoid inactivity.” There’s plenty of evidence to show that aerobic and resistance exercise training improves many cancer-related health outcomes, reducing anxiety, depression, and fatigue while improving physical functioning and quality of life.
Physical activity before and after a diagnosis of cancer also may help to improve survival in some cancers, with perhaps the greatest benefits coming from exercise during and/or after cancer treatment.
Based on the evidence, the panel recommends that cancer survivors engage in moderate-intensity exercise, including aerobic and resistance training, at least two to three times a week. They should exercise for about 30 minutes per session.
The recommendation is based on added confirmation that exercise is generally safe for cancer survivors. The data indicate exercise can lead to improvements in anxiety, depression, fatigue, overall quality of life, and in some cases survival.
The panel also recommends that treatment teams and fitness professionals more systematically incorporate “exercise prescriptions” into cancer care. They should develop the resources to design exercise prescriptions that deliver the right amount of exercise to meet the specific needs, preferences, and abilities of people with cancer.
The ACSM has launched the “Moving Through Cancer” initiative. This initiative will help raise awareness about the importance of exercise during cancer treatment and help support doctors in advising their patients on those benefits.
It’s worth noting that there are still many fascinating questions to explore. While exercise is known to support better health in a variety of ways, correlation is not the same as causation. Questions remain about the underlying mechanisms that may help to explain the observed associations between physical activity, lowered cancer risk, and improved cancer survival.
An intensive NIH research effort, called the Molecular Transducers of Physical Activity Consortium (MoTrPAC), is underway to identify molecular mechanisms that might explain the wide-ranging benefits of physical exercise. It might well shed light on cancer, too.
As that evidence continues to come in, the findings are yet another reminder of the importance of exercise to our health. Everybody—people who are healthy, those with cancer, and cancer survivors alike—should make an extra effort to remain as physically active as our ages, abilities, and current health will allow. If I needed any more motivation to keep up my program of vigorous exercise twice a week, guided by an experienced trainer, here it is!
References:
[1] Exercise Is Medicine in Oncology: Engaging Clinicians to Help Patients Move Through Cancer. Schmitz KH, Campbell AM, Stuiver MM, Pinto BM, Schwartz AL, Morris GS, Ligibel JA, Cheville A, Galvão, DA, Alfano CM, Patel AV, Hue T, Gerber LH, Sallis R, Gusani NJ, Stout NL, Chan L, Flowers F, Doyle C, Helmrich S, Bain W, Sokolof J, Winters-Stone KM, Campbell KL, Matthews CE. CA Cancer J Clin. 2019 Oct 16 [Epub ahead of publication]
[2] American College of Sports Medicine Roundtable Report on Physical Activity, Sedentary Behavior, and Cancer Prevention and Control. Patel AV, Friedenreich CM, Moore SC, Hayes SC, Silver JK, Campbell KL, Gerber LH, George SM, Fulton JE, Denlinger C, Morris GS, Hue T, Schmitz KH, Matthews CE. Med Sci Sports Exerc. 2019 Oct 16. [Epub ahead of publication]
[3] Exercise Guidelines for Cancer Survivors: Consensus Statement from International Multidisciplinary Roundtable. Campbell KL, Winters-Stone KM, Wiskemann J, May AM, Schwartz AL, Courneya KS, Zucker DS, Matthews CE, Ligibel JA, Gerber LH, Morris GS, Patel AV, Hue TF, Perna FM, Schmitz KH. Med Sci Sports Exerc. 2019 Oct 16. [Epub ahead of publication]
Links:
Physical Activity and Cancer (National Cancer Institute/NIH)
Moving Through Cancer (American College of Sports Medicine, Indianapolis, IN)
American College of Sports Medicine
Charles Matthews (NCI)
Frank Perna (NCI)
NIH Support: National Cancer Institute
Posted In: News
Tags: ACSM, American College of Sports Medicine, bladder cancer, breast cancer, cancer, cancer prevention, cancer survivor, colon cancer, endometrial cancer, esophageal cancer, exercise, exercise guidelines, kidney cancer, liver cancer, lung cancer, Molecular Transducers of Physical Activity Consortium, MoTrPAC, Moving Through Cancer, physical activity, physical fitness, stomach cancer
KRAS Targeted Cancer Strategy Shows Early Promise
Posted on February 13th, 2018 by Dr. Francis Collins
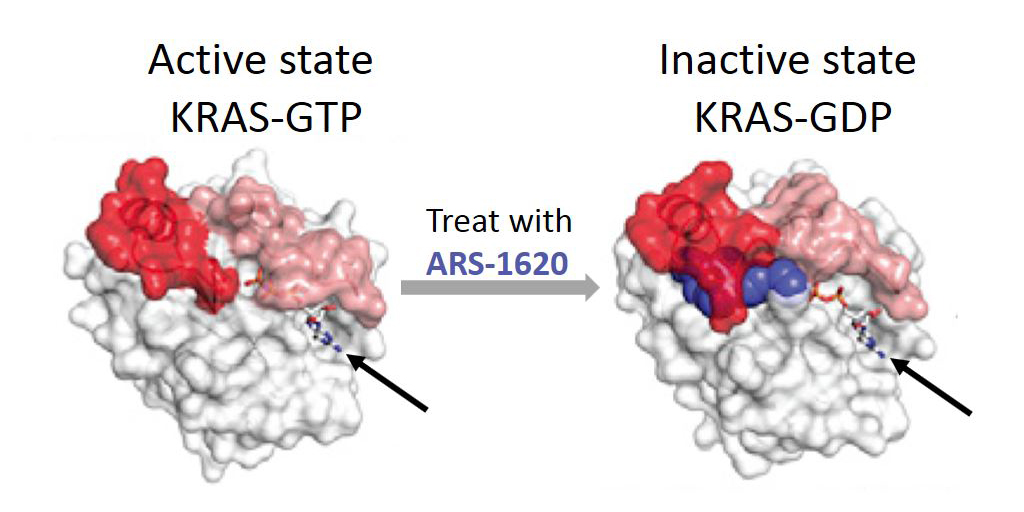
Caption: Mutant KRAS protein (white) keeps switch (red/pink) open in active state for GTP (arrow). After treatment with ARS-1620 (blue), switch is trapped in inactive GDP-bound state.
Credit: Adapted from Cell. 2018 Jan 25;172(3):578-589.
Of the more than 1.7 million Americans expected to be diagnosed with cancer this year, nearly one-third will have tumors that contain at least one mutation in the RAS family of genes [1]. That includes 95 percent of pancreatic cancers and 45 percent of colon cancers. These mutations result in the production of defective proteins that can drive cancer’s uncontrolled growth, as well as make cancers resistant to therapies. As you might expect, RAS has emerged as a major potential target for fighting cancer. Unfortunately, it is a target that’s proven very difficult to “hit” despite nearly three decades of work by researchers in both the private and public sectors, leading NIH’s National Cancer Institute to begin The RAS Initiative in 2013. This important effort has made advances with RAS that have translational potential.
Recently, I was excited to hear of progress in targeting a specific mutant form of KRAS, which is a protein encoded by a _RA_S gene involved in many lung cancers and some pancreatic and colorectal cancers. The new study, carried out by a pharmaceutical research team in mouse models of human cancer, is the first to show that it is possible to shrink a tumor in a living creature by directly inhibiting mutant KRAS protein [2].
Tags: ARS-1620, cancer, colorectal cancer, GTD, GTP, KRAS, lung cancer, non-small cell lung cancer, pancreatic cancer, precision oncology, RAS, small molecules, targeted cancer therapy, The Ras Initiative
New ‘Liquid Biopsy’ Shows Early Promise in Detecting Cancer
Posted on January 30th, 2018 by Dr. Francis Collins
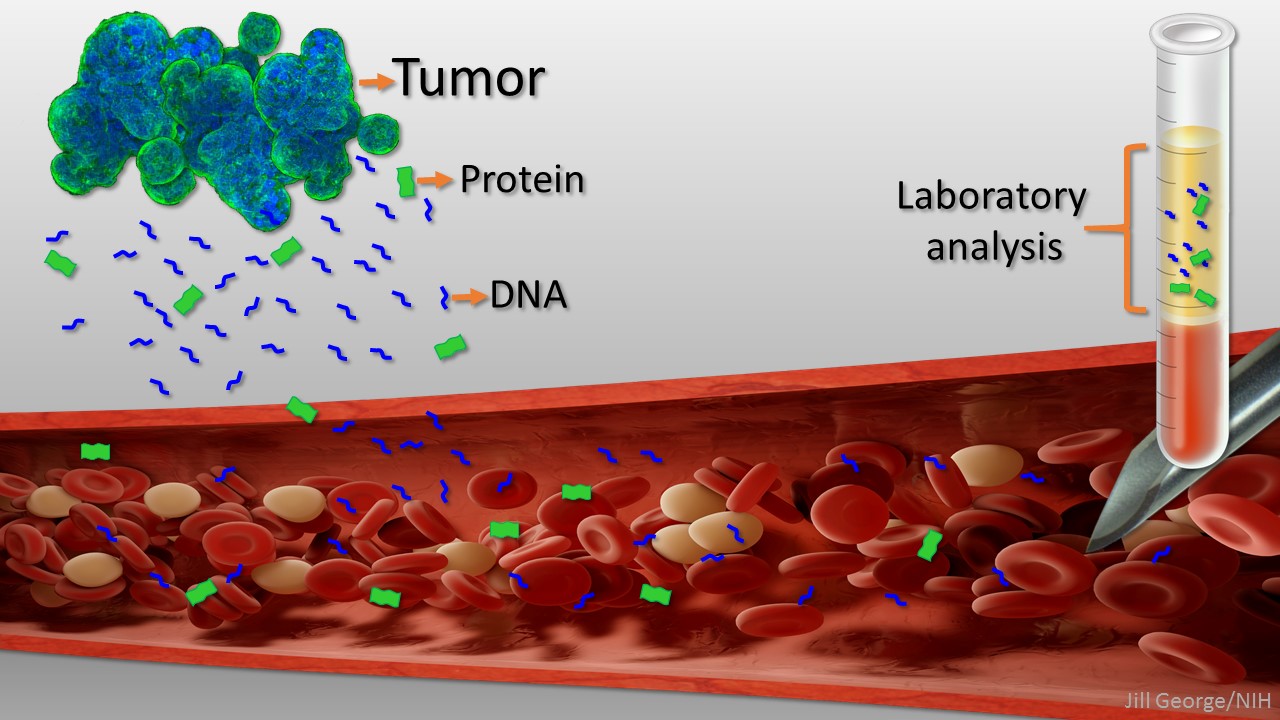
Caption: Liquid biopsy. Tumor cells shed protein and DNA into bloodstream for laboratory analysis and early cancer detection.
Early detection usually offers the best chance to beat cancer. Unfortunately, many tumors aren’t caught until they’ve grown relatively large and spread to other parts of the body. That’s why researchers have worked so tirelessly to develop new and more effective ways of screening for cancer as early as possible. One innovative approach, called “liquid biopsy,” screens for specific molecules that tumors release into the bloodstream.
Recently, an NIH-funded research team reported some encouraging results using a “universal” liquid biopsy called CancerSEEK [1]. By analyzing samples of a person’s blood for eight proteins and segments of 16 genes, CancerSEEK was able to detect most cases of eight different kinds of cancer, including some highly lethal forms—such as pancreatic, ovarian, and liver—that currently lack screening tests.
In a study of 1,005 people known to have one of eight early-stage tumor types, CancerSEEK detected the cancer in blood about 70 percent of the time, which is among the best performances to date for a blood test. Importantly, when CancerSEEK was performed on 812 healthy people without cancer, the test rarely delivered a false-positive result. The test can also be run relatively cheaply, at an estimated cost of less than $500.
Posted In: Health, Science, technology
Tags: blood test, breast cancer, cancer, cancer blood test, cancer detection, cancer diagnostics, CancerSEEK, clinical study, colorectal cancer, early detection, esophageal cancer, liquid biopsy, liver cancer, lung cancer, machine learning, ovarian cancer, pancreatic cancer, stomach cancer, universal liquid biopsy
Random Mutations Play Major Role in Cancer
Posted on April 4th, 2017 by Dr. Francis Collins
 We humans are wired to search for a causative agent when something bad happens. When someone develops cancer, we seek a reason. Maybe cancer runs in the family. Or perhaps the person smoked, never wore sunscreen, or drank too much alcohol. At some level, those are reasonable assumptions, as genes, lifestyle, and environment do play important roles in cancer. But a new study claims that the reason why many people get cancer is simply just bad luck.
We humans are wired to search for a causative agent when something bad happens. When someone develops cancer, we seek a reason. Maybe cancer runs in the family. Or perhaps the person smoked, never wore sunscreen, or drank too much alcohol. At some level, those are reasonable assumptions, as genes, lifestyle, and environment do play important roles in cancer. But a new study claims that the reason why many people get cancer is simply just bad luck.
This bad luck occurs during the normal process of cell division that is essential to helping our bodies grow and remain healthy. Every time a cell divides, its 6 billion letters of DNA are copied, with a new copy going to each daughter cell. Typos inevitably occur during this duplication process, and the cell’s DNA proofreading mechanisms usually catch and correct these typos. However, every once in a while, a typo slips through—and if that misspelling happens to occur in certain key areas of the genome, it can drive a cell onto a pathway of uncontrolled growth that leads to cancer. In fact, according to a team of NIH-funded researchers, nearly two-thirds of DNA typos in human cancers arise in this random way.
The latest findings should help to reassure people being treated for many forms of cancer that they likely couldn’t have prevented their illness. They also serve as an important reminder that, in addition to working on better strategies for prevention, cancer researchers must continue to pursue innovative technologies for early detection and treatment.
Tags: cancer, cancer etiology, cancer incidence, cancer mutations, cancer prevention, Cancer Research UK, cancer risk, cell biology, cell division, DNA, DNA copying errors, DNA sequencing, DNA typos, driver mutations, environment, gene mutations, hereditary, inherited, International Agency for Research on Cancer, lifestyle, lung cancer, mathematics, random mutations, somatic mutations, stem cells, The Cancer Genome Atlas
Are E-cigarettes Leading More Kids to Smoke?
Posted on January 31st, 2017 by Dr. Francis Collins

Thinkstock\MilknCoffee
Today, thanks to decades of educational efforts about the serious health consequences of inhaled tobacco, fewer young people than ever smoke cigarettes in the United States. So, it’s interesting that a growing of number of middle and high school kids are using e-cigarettes—electronic devices that vaporize flavored liquid that generally contains nicotine.
E-cigarettes come with their own health risks, including lung inflammation, asthma, and respiratory infections. But their supporters argue that “vaping,” as it’s often called, might provide an option that would help young people steer clear of traditional cigarettes and the attendant future risks of lung cancer, emphysema, heart disease, and other serious health conditions. Now, a new NIH-funded study finds that this is—pardon the pun—mostly a pipe dream.
Analyzing the self-reported smoking behaviors of thousands of schoolkids nationwide, researchers found no evidence that the availability of e-cigarettes has served to accelerate the decline in youth smoking. In fact, the researchers concluded the opposite: the popularity of e-cigarettes has led more kids—not fewer—to get hooked on nicotine, which meets all criteria for being an addictive substance.
Tags: addiction, adolescence, asthma, behavior, child health, cigarettes, e-cigarettes, electronic cigarettes, emphysema, health education, heart disease, high school, lung cancer, lung inflammation, middle school, National Youth Tobacco Survey, nicotine, pediatrics, respiratory infection, smoking, tobacco, vaping