PMI – NIH Director's Blog (original) (raw)
Cardiometabolic Disease: Big Data Tackles a Big Health Problem
Posted on September 8th, 2016 by Dr. Francis Collins
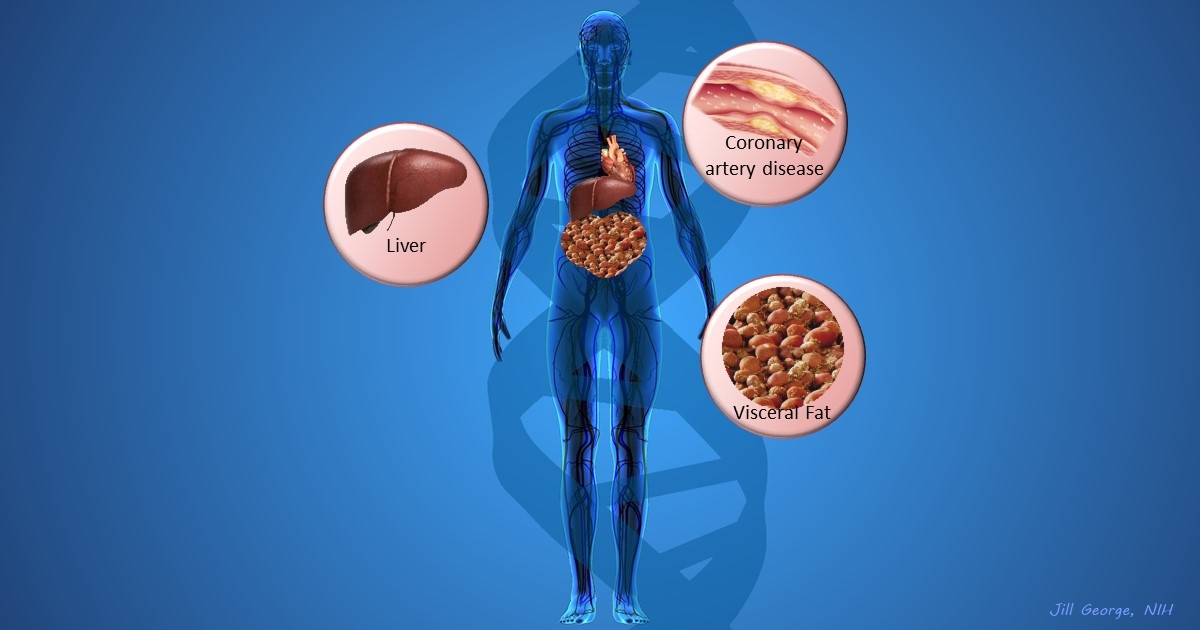
More and more studies are popping up that demonstrate the power of Big Data analyses to get at the underlying molecular pathology of some of our most common diseases. A great example, which may have flown a bit under the radar during the summer holidays, involves cardiometabolic disease. It’s an umbrella term for common vascular and metabolic conditions, including hypertension, impaired glucose and lipid metabolism, excess belly fat, and inflammation. All of these components of cardiometabolic disease can increase a person’s risk for a heart attack or stroke.
In the study, an international research team tapped into the power of genomic data to develop clearer pictures of the complex biocircuitry in seven types of vascular and metabolic tissue known to be affected by cardiometabolic disease: the liver, the heart’s aortic root, visceral abdominal fat, subcutaneous fat, internal mammary artery, skeletal muscle, and blood. The researchers found that while some circuits might regulate the level of gene expression in just one tissue, that’s often not the case. In fact, the researchers’ computational models show that such genetic circuitry can be organized into super networks that work together to influence how multiple tissues carry out fundamental life processes, such as metabolizing glucose or regulating lipid levels. When these networks are perturbed, perhaps by things like inherited variants that affect gene expression, or environmental influences such as a high-carb diet, sedentary lifestyle, the aging process, or infectious disease, the researchers’ modeling work suggests that multiple tissues can be affected, resulting in chronic, systemic disorders including cardiometabolic disease.
Tags: bad cholesterol, big data, bioinformatics, blood lipids, cardiometabolic disease, cardiovascular disease, coronary artery disease, coronary bypass surgery, drug delivery, eQTL, Estonia, fat, gemomics, gene networks, gene variants, GWAS, hyperlipidemia, hypertension, LDL, liver, metabolic syndrome, NHGRI GWAS Catalog, PCSK9, PMI, Precision Medicine Initiative Cohort Program, STARNET, Sweden, systems biology, systems genetics, type 2 diabetes, visceral abdominal fat, visceral fat
Precision Medicine: Using Genomic Data to Predict Drug Side Effects and Benefits
Posted on June 14th, 2016 by Dr. Francis Collins

People with type 2 diabetes are at increased risk for heart attacks, stroke, and other forms of cardiovascular disease, and at an earlier age than other people. Several years ago, the Food and Drug Administration (FDA) recommended that drug developers take special care to show that potential drugs to treat diabetes don’t adversely affect the cardiovascular system [1]. The challenge in implementing that laudable exhortation is that a drug’s long-term health risks may not become clear until thousands or even tens of thousands of people have received it over the course of many years, sometimes even decades.
Now, a large international study, partly funded by NIH, offers some good news: proof-of-principle that “Big Data” tools can help to identify a drug’s potential side effects much earlier in the drug development process [2]. The study, which analyzed vast troves of genomic and clinical data collected over many years from more than 50,000 people with and without diabetes, indicates that anti-diabetes therapies that lower glucose by targeting the product of a specific gene, called GLP1R, are unlikely to boost the risk of cardiovascular disease. In fact, the evidence suggests that such drugs might even offer some protection against heart disease.
Genetic approaches have increasingly been used to identify potentially promising new drug targets. In the study reported in Science Translational Medicine, researchers led by Robert Scott and Nick Wareham from the University of Cambridge, England, and Dawn Waterworth from GlaxoSmithKline, King of Prussia, PA, also wanted to explore whether genomic data could yield important clues about the potential side effects of drugs targeting particular genes.
Tags: anti-diabetes drugs, big data, cardiovascular disease, diabetes, diabetic heart disease, drug benefits, drug development, drug side effects, drug targets, genetic variant, genomics, GlaxoSmithKline, GLP1R, heart disease, investigational drugs, liraglutide, Mendelian randomization, PMI, precision medicine, Precision Medicine Initiative cohort, T2D, type 2 diabetes, Victoza
Happy New Year … and a Look Back at a Memorable 2015
Posted on January 5th, 2016 by Dr. Francis Collins
 A new year has arrived, and it’s going to be an amazing one for biomedical research. But before diving into our first “new science” post of 2016, let’s take a quick look back at 2015 and some of its remarkable accomplishments. A great place to reflect on “the year that was” is the journal Science’s annual Top 10 list of advances in all of scientific research worldwide. Four of 2015’s Top 10 featured developments directly benefited from NIH support—including Science’s “Breakthrough of the Year,” the CRISPR/Cas9 gene-editing technique. Here’s a little more on the NIH-assisted breakthroughs:
A new year has arrived, and it’s going to be an amazing one for biomedical research. But before diving into our first “new science” post of 2016, let’s take a quick look back at 2015 and some of its remarkable accomplishments. A great place to reflect on “the year that was” is the journal Science’s annual Top 10 list of advances in all of scientific research worldwide. Four of 2015’s Top 10 featured developments directly benefited from NIH support—including Science’s “Breakthrough of the Year,” the CRISPR/Cas9 gene-editing technique. Here’s a little more on the NIH-assisted breakthroughs:
CRISPR Makes the Cut: I’ve highlighted CRISPR/Cas9 in several posts. This gene-editing system consists of a short segment of RNA that is attached to an enzyme. The RNA is preprogrammed to find a distinct short sequence of DNA and deliver the enzyme, which acts like a scalpel to slice the sequence out of the genome. It’s fast and pretty precise. Although CRISPR/Cas9 isn’t brand-new—it’s been under development as a gene-editing tool for a few years—Science considered 2015 to be “the year that it broke away from the pack.”
Tags: baker's yeast, biotechnology, brain, CRISPR, CRISPR/Cas9, drug development, Ebola, Ebola vaccine, gene drive, global health, hydrocodone, immune system, immunology, infectious diseases, lymphatic system, neuroscience, opiates, opioid, PMI, Precision Medicine Initiative, rVSV-ZEBOV, Science Breakthrough of the Year, Science's Top 10 Science Advances, thebaine, Top 25 Science Stories of 2015, yeast, ZMapp
Bold Blueprint for Precision Medicine Initiative’s Research Cohort
Posted on September 23rd, 2015 by Dr. Francis Collins
Caption: #PMINetwork Twitter chat with @NIHDirector Francis Collins, NIH Media Branch’s @RenateMyles, and, in background, PMI Cohort Program Acting Director @NCCIH_Josie Briggs.
Credit: @KathyHudsonNIH
Readers of this blog know how excited I am about the potential of precision medicine for revolutionizing efforts to treat disease and improve human health. So, it stands to reason that I’m delighted by the positive reactions of researchers, health professionals, and the public to a much-anticipated report from the Precision Medicine Initiative (PMI) Working Group of the Advisory Committee to the NIH Director. Topping the report’s list of visionary recommendations? Build a national research cohort of 1 million or more Americans over the next three to four years to expand knowledge and practice of precision medicine.
When the President announced PMI during his 2015 State of the Union address, he envisioned a precise new era in medicine in which every patient receives the right treatment at the right time—an era in which health care professionals have the resources at hand to take into account individual differences in genes, environments, and lifestyles that contribute to disease. To achieve this, PMI’s national research cohort would tap into recent advances in science, technology, and research participation policies to build the knowledge base needed to develop individualized care for all diseases and conditions.
Tags: cohort, Obama's Precision Medicine plan, PMI, PMI national research cohort, PMI research cohort, PMI Working Group, precision medicine, Precision Medicine Initiative, Precision Medicine Initiative cohort, Precision Medicine Working Group, Twitter chat
