respiratory diseases – NIH Director's Blog (original) (raw)
Clinical Center Doctors Testing 3D-Printed Miniature Ventilator
Posted on November 29th, 2022 by James K. Gilman, MD, NIH Clinical Center

Caption: A USB flash drive (front) next to the 3D-printed miniature ventilator (back). Credit: William Pritchard, Clinical Center, NIH
Here at the NIH Clinical Center, we are proud to be considered a world-renowned research hospital that provides hope through pioneering clinical research to improve human health. But what you may not know is that our doctors are constantly partnering with public and private sectors to come up with innovative technologies that will help to advance health outcomes.
I’m excited to bring to you a story that is perfect example of the ingenuity of our NIH doctors working with global strategic partners to create potentially life-saving technologies. This story begins during the COVID-19 pandemic with the global shortage of ventilators to help patients breathe. Hospitals had a profound need for inexpensive, easy-to-use, rapidly mass-produced resuscitation devices that could be quickly distributed in areas of critical need.
Through strategic partnerships, our Clinical Center doctors learned about and joined an international group of engineers, physicians, respiratory therapists, and patient advocates using their engineering skills to create a ventilator that was functional, affordable, and intuitive. After several iterations and bench testing, they devised a user-friendly ventilator.
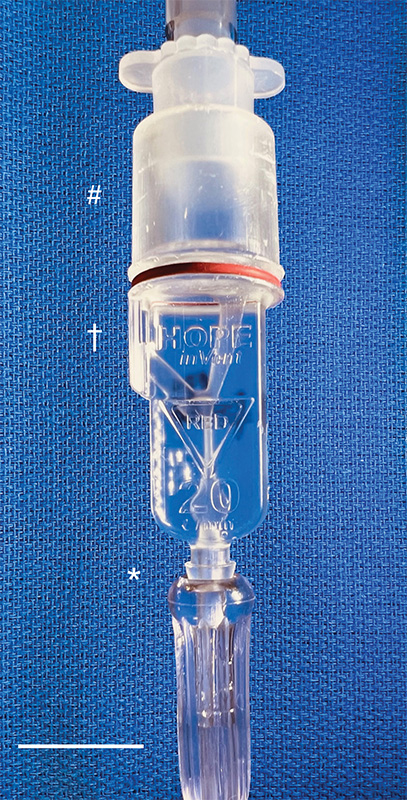
Caption: The miniature ventilator connected to an oxygen line (asterisk) and the breathing tube to the patient (crosshatch). The exhaust (dagger) is recessed to prevent accidental blockage. Credit: William Pritchard, Clinical Center, NIH
Then, with the assistance of 3D-printing technology, they improved the original design and did something pretty incredible: the team created the smallest single-patient ventilator seen to date. The device is just 2.4 centimeters (about 1 inch) in diameter with a length of 7.4 centimeters (about 3 inches).
A typical ventilator in a hospital obviously is much larger and has a bellows system. It fills with oxygen and then forces it into the lungs followed by the patient passively exhaling. These systems have multiple moving parts, valves, hoses, and electronic or mechanical controls to manage all aspects of the oxygen flow into the lungs.
But our miniature, 3D-printed ventilator is single use, disposable, and has no moving parts. It’s based on principles of fluidics to ventilate patients by automatically oscillating between forced inspiration and assisted expiration as airway pressure changes. It requires only a continuous supply of pressurized oxygen.
The possibilities of this 3D-printed miniature ventilator are broad. The ventilators could be easily used in emergency transport, potentially treating battlefield casualties or responding to disasters and mass casualty events like earthquakes.
While refining a concept is important, the key is converting it to actual use, which our doctors are doing admirably in their preclinical and clinical studies. NIH’s William Pritchard, Andrew Mannes, Brad Wood, John Karanian, Ivane Bakhutashvili, Matthew Starost, David Eckstein, and medical student Sheridan Reed studied and have already tested the ventilators in swine with acute lung injury, a common severe outcome in a number of respiratory threats including COVID-19.
In the study, the doctors tested three versions of the device built to correspond to mild, moderate, and severe lung injury. The respirators provided adequate support for moderate and mild lung injuries, and the doctors recall how amazing it was initially to witness a 190-pound swine ventilated by this miniature ventilator.
The doctors believe that the 3D-printed miniature ventilator is a potential “game changer” from start to finish since it is lifesaving, small, simple to use, can be easily and inexpensively printed and stored, and does not require additional maintenance. They recently published their preclinical trial results in the journal Science Translational Medicine [1].
The NIH team is preparing to initiate first-in-human trials here at the Clinical Center in the coming months. Perhaps, in the not-too-distant future, a device designed to help people breathe could fit into your pocket next to your phone and keys.
Reference:
[1] In-line miniature 3D-printed pressure-cycled ventilator maintains respiratory homeostasis in swine with induced acute pulmonary injury. Pritchard WF, Karanian JW, Jung C, Bakhutashvili I, Reed SL, Starost MF, Froelke BR, Barnes TR, Stevenson D, Mendoza A, Eckstein DJ, Wood BJ, Walsh BK, Mannes AJ. Sci Transl Med. 2022 Oct 12;14(666):eabm8351.
Links:
Clinical Center (NIH)
Andrew Mannes (Clinical Center)
Bradford Wood (Clinical Center)
David Eckstein (Clinical Center)
Note: Dr. Lawrence Tabak, who performs the duties of the NIH Director, has asked the heads of NIH’s Institutes and Centers (ICs) to contribute occasional guest posts to the blog to highlight some of the interesting science that they support and conduct. This is the 21st in the series of NIH IC guest posts that will run until a new permanent NIH director is in place.
Posted In: Generic
Tags: 3D printing, battlefield, biomedical engineering, breathing, breathing problems, clinical research, clinical trial, COVID-19, disaster response, disposable, emergency medicine, emergency transport, hospital, lungs, microfluidics, miniature ventilator, NIH Clinical Center, pandemic, pre-clinical research, pressurized oxygen, respiratory diseases, resuscitation device, single-use, swine, technology, ventilator
Mapping Severe COVID-19 in the Lungs at Single-Cell Resolution
Posted on April 13th, 2021 by Dr. Francis Collins

Caption: Image shows macrophages (red), fibroblast cells (green), and other cells (blue). In late COVID-19, macrophages migrate near fibroblasts, which may play a role in fibrosis. Credit: Images courtesy of André Rendeiro
A crucial question for COVID-19 researchers is what causes progression of the initial infection, leading to life-threatening respiratory illness. A good place to look for clues is in the lungs of those COVID-19 patients who’ve tragically lost their lives to acute respiratory distress syndrome (ARDS), in which fluid and cellular infiltrates build up in the lung’s air sacs, called alveoli, keeping them from exchanging oxygen with the bloodstream.
As shown above, a team of NIH-funded researchers has done just that, capturing changes in the lungs over the course of a COVID-19 infection at unprecedented, single-cell resolution. These imaging data add evidence that SARS-CoV-2, the coronavirus that causes COVID-19, primarily infects cells at the surface of the air sacs. Their findings also offer valuable clues for treating the most severe consequences of COVID-19, suggesting that a certain type of scavenging immune cell might be driving the widespread lung inflammation that leads to ARDS.
The findings, published in Nature [1], come from Olivier Elemento and Robert E. Schwartz, Weill Cornell Medicine, New York. They already knew from earlier COVID-19 studies about the body’s own immune response causing the lung inflammation that leads to ARDS. What was missing was an understanding of the precise interplay between immune cells and lung tissue infected with SARS-CoV-2. It also wasn’t clear how the ARDS seen with COVID-19 compared to the ARDS seen in other serious respiratory diseases, including influenza and bacterial pneumonia.
Traditional tissue analysis uses chemical stains or tagged antibodies to label certain proteins and visualize important features in autopsied human tissues. But using these older techniques, it isn’t possible to capture more than a few such proteins at once. To get a more finely detailed view, the researchers used a more advanced technology called imaging mass cytometry [2].
This approach uses a collection of lanthanide metal-tagged antibodies to label simultaneously dozens of molecular markers on cells within tissues. Next, a special laser scans the labeled tissue sections, which vaporizes the heavy metal tags. As the metals are vaporized, their distinct signatures are detected in a mass spectrometer along with their spatial position relative to the laser. The technique makes it possible to map precisely where a diversity of distinct cell types is located in a tissue sample with respect to one another.
In the new study, the researchers applied the method to 19 lung tissue samples from patients who had died of severe COVID-19, acute bacterial pneumonia, or bacterial or influenza-related ARDS. They included 36 markers to differentiate various types of lung and immune cells as well as the SARS-CoV-2 spike protein and molecular signs of immune activation, inflammation, and cell death. For comparison, they also mapped four lung tissue samples from people who had died without lung disease.
Altogether, they captured more than 200 lung tissue maps, representing more than 660,000 cells across all the tissues sampled. Those images showed in all cases that respiratory infection led to a thickening of the walls surrounding alveoli as immune cells entered. They also showed an increase in cell death in infected compared to healthy lungs.
Their maps suggest that what happens in the lungs of COVID-19 patients who die with ARDS isn’t entirely unique. It’s similar to what happens in the lungs of those with other life-threatening respiratory infections who also die with ARDS.
They did, however, reveal a potentially prominent role in COVID-19 for white blood cells called macrophages. The results showed that macrophages are much more abundant in the lungs of severe COVID-19 patients compared to other lung infections.
In late COVID-19, macrophages also increase in the walls of alveoli, where they interact with lung cells known as fibroblasts. This suggests these interactions may play a role in the buildup of damaging fibrous tissue, or scarring, in the alveoli that tends to be seen in severe COVID-19 respiratory infections.
While the virus initiates this life-threatening damage, its progression may not depend on the persistence of the virus, but on an overreaction of the immune system. This may explain why immunomodulatory treatments like dexamethasone can provide benefit to the sickest patients with COVID-19. To learn even more, the researchers are making their data and maps available as a resource for scientists around the world who are busily working to understand this devastating illness and help put an end to the terrible toll caused by this pandemic.
References:
[1] The spatial landscape of lung pathology during COVID-19 progression. Rendeiro AF, Ravichandran H, Bram Y, Chandar V, Kim J, Meydan C, Park J, Foox J, Hether T, Warren S, Kim Y, Reeves J, Salvatore S, Mason CE, Swanson EC, Borczuk AC, Elemento O, Schwartz RE. Nature. 2021 Mar 29.
[2] Mass cytometry imaging for the study of human diseases-applications and data analysis strategies. Baharlou H, Canete NP, Cunningham AL, Harman AN, Patrick E. Front Immunol. 2019 Nov 14;10:2657.
Links:
COVID-19 Research (NIH)
Elemento Lab (Weill Cornell Medicine, New York)
Schwartz Lab (Weill Cornell Medicine)
NIH Support: National Center for Advancing Translational Sciences; National Institute of Allergy and Infectious Diseases; National Institute of Diabetes and Digestive and Kidney Diseases; National Cancer Institute
Posted In: News
Tags: acute respiratory distress syndrome, alveoli, ARDS, bacterial pneumonia, COVID-19, COVID-19 immune response, dexamethasone, fibroblasts, fibrosis, imaging mass cytometry, lung inflammation, lung pathology, lung tissue map, lungs, macrophages, novel coronavirus, pandemic, respiratory diseases, SARS-CoV-2, single-cell resolution, spike protein
Months After Recovery, COVID-19 Survivors Often Have Persistent Lung Trouble
Posted on September 15th, 2020 by Dr. Francis Collins

Caption: Testing breathing capacity with a spirometer. Credit: iStock/Koldunov
The pandemic has already claimed far too many lives in the United States and around the world. Fortunately, as doctors have gained more experience in treating coronavirus disease 2019 (COVID-19), more people who’ve been hospitalized eventually will recover. This raises an important question: what does recovery look like for them?
Because COVID-19 is still a new condition, there aren’t a lot of data out there yet to answer that question. But a recent study of 55 people recovering from COVID-19 in China offers some early insight into the recovery of lung function [1]. The results make clear that—even in those with a mild-to-moderate infection—the effects of COVID-19 can persist in the lungs for months. In fact, three months after leaving the hospital about 70 percent of those in the study continued to have abnormal lung scans, an indication that the lungs are still damaged and trying to heal.
The findings in EClinicalMedicine come from a team in Henan Province, China, led by Aiguo Xu, The First Affiliated Hospital of Zhengzhou University; Yanfeng Gao, Zhengzhou University; and Hong Luo, Guangshan People’s Hospital. They’d heard about reports of lung abnormalities in patients discharged from the hospital. But it wasn’t clear how long those problems stuck around.
To find out, the researchers enrolled 55 men and women who’d been admitted to the hospital with COVID-19 three months earlier. Some of the participants, whose average age was 48, had other health conditions, such as diabetes or heart disease. But none had any pre-existing lung problems.
Most of the patients had mild or moderate respiratory illness while hospitalized. Only four of the 55 had been classified as severely ill. Fourteen patients required supplemental oxygen while in the hospital, but none needed mechanical ventilation.
Three months after discharge from the hospital, all of the patients were able to return to work. But they continued to have lingering symptoms of COVID-19, including shortness of breath, cough, gastrointestinal problems, headache, or fatigue.
Evidence of this continued trouble also showed up in their lungs. Thirty-nine of the study’s participants had an abnormal result in their computed tomography (CT) lung scan, which creates cross-sectional images of the lungs. Fourteen individuals (1 in 4) also showed reduced lung function in breathing tests.
Interestingly, the researchers found that those who went on to have more lasting lung problems also had elevated levels of D-dimer, a protein fragment that arises when a blood clot dissolves. They suggest that a D-dimer test might help to identify those with COVID-19 who would benefit from pulmonary rehabilitation to rebuild their lung function, even in the absence of severe respiratory symptoms.
This finding also points to the way in which the SARS-CoV-2 virus seems to enhance a tendency toward blood clotting—a problem addressed in our Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) public-private partnership. The partnership recently initiated a trial of blood thinners. That trial will start out by focusing on newly diagnosed outpatients and hospitalized patients, but will go on to include a component related to convalescence.
Moving forward, it will be important to conduct larger and longer-term studies of COVID-19 recovery in people of diverse backgrounds to continue to learn more about what it means to survive COVID-19. The new findings certainly indicate that for many people who’ve been hospitalized with COVID-19, regaining normal lung function may take a while. As we learn even more about the underlying causes and long-term consequences of this new infectious disease, let’s hope it will soon lead to insights that will help many more COVID-19 long-haulers and their concerned loved ones breathe easier.
Reference:
[1] Follow-up study of the pulmonary function and related physiological characteristics of COVID-19 survivors three months after recovery. Zhao YM, Shang YM, Song WB, Li QQ, Xie H, Xu QF, Jia JL, Li LM, Mao HL, Zhou XM, Luo H, Gao YF, Xu AG. EClinicalMedicine.2020 Aug 25:100463
Links:
Coronavirus (COVID-19) (NIH)
How the Lungs Work (National Heart, Lung, and Blood Institute/NIH)
Computed Tomography (CT) (National Institute of Biomedical Imaging and Bioengineering/NIH)
Zhengzhou University (Zhengzhou City, Henan Province, China)
Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) (NIH)
Posted In: News
Tags: ACTIV, blood, blood clots, blood thinner, China, computed tomography, coronavirus, COVID-19, COVID-19 pulmonary disease, COVID-19 recovery, COVID-19 survivor, CT scan, D-dimer, long COVID, long-haulers, lung scan, lungs, pandemic, pulmonary disease, respiratory diseases, SARS-CoV-2
Structural Biology Points Way to Coronavirus Vaccine
Posted on March 3rd, 2020 by Dr. Francis Collins
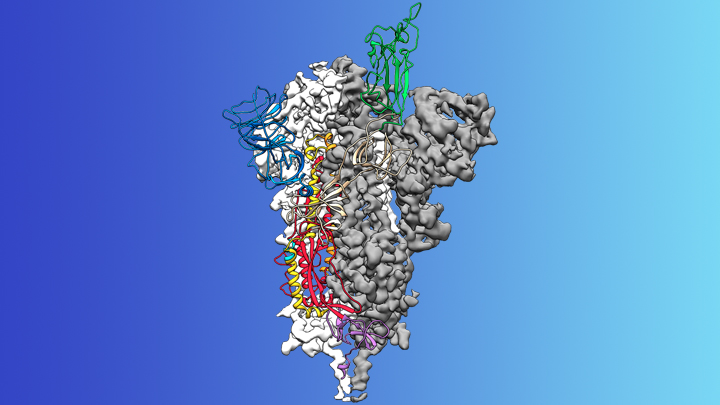
Caption: Atomic-level structure of the spike protein of the virus that causes COVID-19.
Credit: McLellan Lab, University of Texas at Austin
The recent COVID-19 outbreak of a novel type of coronavirus that began in China has prompted a massive global effort to contain and slow its spread. Despite those efforts, over the last month the virus has begun circulating outside of China in multiple countries and territories.
Cases have now appeared in the United States involving some affected individuals who haven’t traveled recently outside the country. They also have had no known contact with others who have recently arrived from China or other countries where the virus is spreading. The NIH and other U.S. public health agencies stand on high alert and have mobilized needed resources to help not only in its containment, but in the development of life-saving interventions.
On the treatment and prevention front, some encouraging news was recently reported. In record time, an NIH-funded team of researchers has created the first atomic-scale map of a promising protein target for vaccine development [1]. This is the so-called spike protein on the new coronavirus that causes COVID-19. As shown above, a portion of this spiky surface appendage (green) allows the virus to bind a receptor on human cells, causing other portions of the spike to fuse the viral and human cell membranes. This process is needed for the virus to gain entry into cells and infect them.
Preclinical studies in mice of a candidate vaccine based on this spike protein are already underway at NIH’s Vaccine Research Center (VRC), part of the National Institute of Allergy and Infectious Diseases (NIAID). An early-stage phase I clinical trial of this vaccine in people is expected to begin within weeks. But there will be many more steps after that to test safety and efficacy, and then to scale up to produce millions of doses. Even though this timetable will potentially break all previous speed records, a safe and effective vaccine will take at least another year to be ready for widespread deployment.
Coronaviruses are a large family of viruses, including some that cause “the common cold” in healthy humans. In fact, these viruses are found throughout the world and account for up to 30 percent of upper respiratory tract infections in adults.
This outbreak of COVID-19 marks the third time in recent years that a coronavirus has emerged to cause severe disease and death in some people. Earlier coronavirus outbreaks included SARS (severe acute respiratory syndrome), which emerged in late 2002 and disappeared two years later, and MERS (Middle East respiratory syndrome), which emerged in 2012 and continues to affect people in small numbers.
Soon after COVID-19 emerged, the new coronavirus, which is closely related to SARS, was recognized as its cause. NIH-funded researchers including Jason McLellan, an alumnus of the VRC and now at The University of Texas at Austin, were ready. They’d been studying coronaviruses in collaboration with NIAID investigators for years, with special attention to the spike proteins.
Just two weeks after Chinese scientists reported the first genome sequence of the virus [2], McLellan and his colleagues designed and produced samples of its spike protein. Importantly, his team had earlier developed a method to lock coronavirus spike proteins into a shape that makes them both easier to analyze structurally via the high-resolution imaging tool cryo-electron microscopy and to use in vaccine development efforts.
After locking the spike protein in the shape it takes before fusing with a human cell to infect it, the researchers reconstructed its atomic-scale 3D structural map in just 12 days. Their results, published in Science, confirm that the spike protein on the virus that causes COVID-19 is quite similar to that of its close relative, the SARS virus. It also appears to bind human cells more tightly than the SARS virus, which may help to explain why the new coronavirus appears to spread more easily from person to person, mainly by respiratory transmission.
McLellan’s team and his NIAID VRC counterparts also plan to use the stabilized spike protein as a probe to isolate naturally produced antibodies from people who’ve recovered from COVID-19. Such antibodies might form the basis of a treatment for people who’ve been exposed to the virus, such as health care workers.
The NIAID is now working with the biotechnology company Moderna, Cambridge, MA, to use the latest findings to develop a vaccine candidate using messenger RNA (mRNA), molecules that serve as templates for making proteins. The goal is to direct the body to produce a spike protein in such a way to elicit an immune response and the production of antibodies. An early clinical trial of the vaccine in people is expected to begin in the coming weeks. Other vaccine candidates are also in preclinical development.
Meanwhile, the first clinical trial in the U.S. to evaluate an experimental treatment for COVID-19 is already underway at the University of Nebraska Medical Center’s biocontainment unit [3]. The NIH-sponsored trial will evaluate the safety and efficacy of the experimental antiviral drug remdesivir in hospitalized adults diagnosed with COVID-19. The first participant is an American who was repatriated after being quarantined on the Diamond Princess cruise ship in Japan.
As noted, the risk of contracting COVID-19 in the United States is currently low, but the situation is changing rapidly. One of the features that makes the virus so challenging to stay in front of is its long latency period before the characteristic flu-like fever, cough, and shortness of breath manifest. In fact, people infected with the virus may not show any symptoms for up to two weeks, allowing them to pass it on to others in the meantime. You can track the reported cases in the United States on the Centers for Disease Control and Prevention’s website.
As the outbreak continues over the coming weeks and months, you can be certain that NIH and other U.S. public health organizations are working at full speed to understand this virus and to develop better diagnostics, treatments, and vaccines.
References:
[1] Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Science. 2020 Feb 19.
[2] A new coronavirus associated with human respiratory disease in China. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. Nature. 2020 Feb 3.
[3] NIH clinical trial of remdesivir to treat COVID-19 begins. NIH News Release. Feb 25, 2020.
Links:
Coronaviruses (National Institute of Allergy and Infectious Diseases/NIH)
Coronavirus (COVID-19) (NIAID)
Coronavirus Disease 2019 (Centers for Disease Control and Prevention, Atlanta)
NIH Support: National Institute of Allergy and Infectious Diseases
Posted In: News
Tags: antibody treatment, China, clinical trials, community spread, coronavirus, COVID-19, cryo-EM, epidemic, infectious disease, MERS, mRNA, mRNA vaccine, novel coronavirus, pandemic, remdesivir, respiratory diseases, SARS, spike protein, structural biology, vaccine, vaccine development, Vaccine Research Center, virology, virus
Creative Minds: Exploring the Health Effects of Fracking
Posted on March 10th, 2016 by Dr. Francis Collins

Elaine Hill
A few years ago, Elaine Hill was a doctoral student in applied economics at Cornell University in Ithaca, NY, studying maize markets in Uganda [1] and dairy supply chains in the northeastern U.S [2]. But when fracking—a controversial, hydraulic fracturing technique used to produce oil and natural gas—became a hot topic in the Finger Lakes region of upstate New York, Hill was motivated to shift gears.
After watching a documentary about fracking, Hill decided to search for scientific evidence on its possible health effects, but found relatively little high-quality data. So, she embarked on a new project—one that eventually earned her a Ph.D.—to evaluate what, if any, impact fracking has on infant and child health. Now, supported by a 2015 NIH Director’s Early Independence Award, Hill is pursuing this line of research further as an assistant professor of Public Health Sciences at the University of Rochester School of Medicine and Dentistry, Rochester, NY.
Tags: 2015 NIH Director’s Early Independence Award, child health, drilling, environmental health, epidemiology, fracking, fracking leases, gas, groundwater, high-pressure hydraulic fracturing, hydraulic fracturing, infant health, International Society for Environmental Epidemiology, oil, pediatrics, reproductive health, respiratory diseases, shale, shale gas well
Gene Expression Test Aims to Reduce Antibiotic Overuse
Posted on February 2nd, 2016 by Dr. Francis Collins

Caption: Duke physician-scientist Ephraim Tsalik assesses a patient for a respiratory infection.
Credit: Shawn Rocco/Duke Health
Without doubt, antibiotic drugs have saved hundreds of millions of lives from bacterial infections that would have otherwise been fatal. But their inappropriate use has led to the rise of antibiotic-resistant superbugs, which now infect at least 2 million Americans every year and are responsible for thousands of deaths [1]. I’ve just come from the World Economic Forum in Davos, Switzerland, where concerns about antibiotic resistance and overuse was a topic of conversation. In fact, some of the world’s biggest pharmaceutical companies issued a joint declaration at the forum, calling on governments and industry to work together to combat this growing public health threat [2].
Many people who go to the doctor suffering from respiratory symptoms expect to be given a prescription for antibiotics. Not only do such antibiotics often fail to help, they serve to fuel the development of antibiotic-resistant superbugs [3]. That’s because antibiotics are only useful in treating respiratory illnesses caused by bacteria, and have no impact on those caused by viruses (which are frequent in the wintertime). So, I’m pleased to report that a research team, partially supported by NIH, recently made progress toward a simple blood test that analyzes patterns of gene expression to determine if a patient’s respiratory symptoms likely stem from a bacterial infection, viral infection, or no infection at all.
In contrast to standard tests that look for signs of a specific infectious agent—respiratory syncytial virus (RSV) or the influenza virus, for instance—the new strategy casts a wide net that takes into account changes in the patterns of gene expression in the bloodstream, which differ depending on whether a person is fighting off a bacterial or a viral infection. As reported in Science Translational Medicine [4], Geoffrey Ginsburg, Christopher Woods, and Ephraim Tsalik of Duke University’s Center for Applied Genomics and Precision Medicine, Durham, NC, and their colleagues collected blood samples from 273 people who came to the emergency room (ER) with signs of acute respiratory illness. Standard diagnostic tests showed that 70 patients arrived in the ER with bacterial infections and 115 were battling viruses. Another 88 patients had no signs of infection, with symptoms traced instead to other health conditions.
Tags: acute respiratory illness, Antibacterial Resistance Leadership Group, antibiotic overuse, antibiotic resistance, antibiotic-resistant infections, antibiotics, applied genomics, bacteria, bacterial infections, blood test, diagnostics, gene expression signature, gene signature, genomics, immunology, infectious disease, noninfectious respiratory problems, precision medicine, procalcitonin, respiratory bacteria, respiratory diseases, respiratory viruses, superbugs, Task Force for Combating Antibiotic-Resistant Bacteria, virology, viruses
Cystic Fibrosis: Keeping the Momentum Going
Posted on December 8th, 2015 by Dr. Francis Collins
Caption: Lower left, me, Lap-Chee Tsui, and John Riordan celebrating our discovery of the cystic fibrosis gene. Right, Robert J. Beall, me, and Doris Tulcin at a November Cystic Fibrosis Foundation event honoring Dr. Beall.
It’s been more than a quarter-century since my colleagues and I were able to identify the gene responsible for cystic fibrosis (CF), a life-shortening inherited disease that mainly affects the lungs and pancreas [1]. And, at a recent event in New York, I had an opportunity to celebrate how far we’ve come since then in treating CF, as well as to honor a major force behind that progress, Dr. Bob Beall, who has just retired as president and chief executive officer of the Cystic Fibrosis Foundation.
Thanks to the tireless efforts of Bob and many others in the public and private sectors to support basic, translational, and clinical research, we today have two therapies from Vertex Pharmaceuticals that are targeted specifically at CF’s underlying molecular cause: ivacaftor (Kalydeco™), approved by the Food and Drug Administration (FDA) in 2012 for people with an uncommon mutation in the CF gene; and the combination ivacaftor-lumacaftor (Orkambi™), approved by the FDA in July for the roughly 50 percent of CF patients with two copies of the most common mutation. Yet more remains to be done before we can truly declare victory. Not only are new therapies needed for people with other CF mutations, but also for those with the common mutation who don’t respond well to Orkambi™. So, the work needs to go on, and I’m encouraged by new findings that suggest a different strategy for helping folks with the most common CF mutation.
Tags: Bob Beall, CF, CFTR, chronic infections, cystic fibrosis, Cystic Fibrosis Foundation, cystic fibrosis transmembrane conductance regulator gene, Doris Tulcin, F508del, genetic disorder, interactome, interactome remodelling, ion channel, ivacaftor, John Riordan, Kalydeco, Lap-Chee Tsui, lumacaftor, lung infections, lungs, misfolded proteins, Orkambi, pancreas, protein networks, proteomics, respiratory diseases, Vertex Pharmaceuticals
Protecting Kids: Developing a Vaccine for Respiratory Syncytial Virus
Posted on November 17th, 2015 by Dr. Francis Collins
 Vaccines are one of biomedicine’s most powerful and successful tools for protecting against infectious diseases. While we currently have safe and effective vaccines to prevent measles, mumps, and a great many other common childhood diseases, we still lack a vaccine to guard against respiratory syncytial virus (RSV)—a leading cause of pneumonia among infants and young children.
Vaccines are one of biomedicine’s most powerful and successful tools for protecting against infectious diseases. While we currently have safe and effective vaccines to prevent measles, mumps, and a great many other common childhood diseases, we still lack a vaccine to guard against respiratory syncytial virus (RSV)—a leading cause of pneumonia among infants and young children.
Each year, more than 2 million U.S. children under the age of 5 require medical care for pneumonia and other potentially life-threatening lower respiratory infections caused by RSV [1,2]. Worldwide, the situation is even worse, with more than 30 million infections estimated to occur annually, most among kids in developing countries, where as many as 200,000 deaths may result [3]. So, I’m pleased to report some significant progress in biomedical research’s long battle against RSV: encouraging early results from a clinical trial of an experimental vaccine specifically designed to outwit the virus.
Tags: childhood disease, childhood infectious diseases, childhood vaccine, clinical trial, CRADA, genetic engineering, global health, immunity, live vaccine, M2-2 gene, neutralizing antibodies, pneumonia, respiratory diseases, respiratory syncytial virus, RSV, RSV MEDI ΔM2-2, RSV vaccine, translational medicine, vaccine, virology
Creative Minds: Harnessing Technologies to Study Air Pollution’s Health Risks
Posted on May 21st, 2015 by Dr. Francis Collins
Perry Hystad
Credit: Hannah O’Leary, Oregon State University
After college, Perry Hystad took a trip to India and, while touring several large cities, noticed the vast clouds of exhaust from vehicles, smoke from factories, and soot from biomass-burning cook stoves. As he watched the rapid urban expansion all around him, Hystad remembers thinking: What effect does breathing such pollution day in and day out have upon these people’s health?
This question stuck with Hystad, and he soon developed a profound interest in environmental health. In 2013, Hystad completed his Ph.D. in his native Canada, studying the environmental risk factors for lung cancer [1, 2, 3]. Now, with the support of an NIH Director’s Early Independence Award, Hystad has launched his own lab at Oregon State University, Corvallis, to investigate further the health impacts of air pollution, which one recent analysis indicates may contribute to as many as several million deaths worldwide each year [4].
Tags: air pollution, air quality, asthma, Bangladesh, biomass, Canada, cardiopulmonary health, clean air, cookstove, environmental health, fine particulate matter, fossil fuels, global health, India, lungs, NIH Director's Early Independence Award, PURE, PURE-AIR, respiratory diseases, satellite technology, South Africa, wearable sensors
Ferreting Out Genomic Secrets
Posted on January 8th, 2015 by Dr. Francis Collins
Ferret in a Colorado conservation center, U.S. Fish and Wildlife Service
Not only is the ferret (Mustela putorius furo) adept at navigating a dirt field or threading electrical cables through piping (in New Zealand, ferrets can be registered as electrician assistants), this furry 5-pounder ranks as a real heavyweight for studying respiratory diseases. In fact, much of our current thinking about influenza is influenced by research with ferrets.
Now, the ferret will stand out even more. As reported online in Nature Biotechnology, NIH-funded researchers recently sequenced the genome of the sable ferret, the type that is bred in the United States as a pet. By studying this genetic blueprint like an explorer would a map, scientists can perform experiments to learn more systematically how the ferret copes biologically with common or emerging respiratory pathogens, pointing the way to improved strategies to preserve the health and well being of humans and ferrets alike.
Tags: animal models, cancer, cystic fibrosis, development, ferret, genome, immune response, respiratory diseases, respiratory viruses, sequencing


