Selected Approaches Ranking Contextual Term for the BioASQ Multi-label Classification (Task6a and 7a) (original) (raw)
Abstract
MeSH annotations are attached to the Medline abstracts to improve retrieval and this service is provided from the curators at the National Library of Medicine (NLM). Efforts to automatically assign such headings to Medline abstracts have proven difficult, on the other side, such approaches would increase throughput and efficiency. Trained solutions, i.e. machine learning solutions, achieve promising results, however these advancements do not fully explain, which features from the text would suit best the identification of MeSH Headings from the abstracts. This manuscript describes new approaches for the identification of contextual features for automatic MeSH annotations, which is a Multi-Label Classification (BioASQ Task6a): more specifically, different approaches for the identification of compound terms have been tested and evaluated. The described system has then been extended to better rank selected labels and has been tested in the BioASQ Task7a challenge. The tests show that our recall measures (see Task6a) have improved and in the second challenge, both the performance for precision and recall were boosted. Our work improves our understanding how contextual features from the text help reduce the performance gap given between purely trained solutions and feature-based solutions (possibly including trained solutions). In addition, we have to point out that the lexical features given from the MeSH thesaurus come with a significant and high discrepancy towards the actual annotations of MeSH Headings attributed by human curators, which also hinders improvements to the automatic annotation of Medline abstracts with MeSH Headings.
Similar content being viewed by others
Keywords
1 Introduction
The scientific biomedical literature is being collected and archived by the National Library of Medicine (NLM) over the past 150 years. Documents have manually been annotated with Medical Subject HeadingsFootnote 1 in order to search and access the documents efficiently. The process of manually assigning indexing terms is very time consuming and thus tedious work. Furthermore, the biomedical literature in PubMed has grown from 12 Million citations in 2004 [4] to 29 Million citations in 2019Footnote 2 having a growth rate of 4% per year [[23](#ref-CR23 "Lu, Z.: PubMed and beyond: a survey of web tools for searching biomedical literature. Database (Oxford), p. baq036 (2011). https://doi.org/10.1093/database/baq036
")\] leading to high pressure in delivering the MeSH annotations.The growth in published biomedical literature as well as the difficulties in manually assigning indexing terms shows the need for routines that automatically annotate and index the scientific articles in order to use metadata terms for information retrieval purposes. At best such supporting automatic solutions should also contribute clues to the curators about the selection of most relevant and best supported terms throughout all the stages of their work. Such clues could be difficult to derive, e.g., from the scientific text, since the MeSH Headings cover mostly compound terms, which – at best – have complex representations in the text.
The Medical Text Indexer (MTI) has been developed by the NLM to provide an automated indexing system for the Medical Subject Headings to the curators. From 2000 onward, the NLM indexing initiative has been initiated, in particular due to the availability of the electronic versions of the scientific articles since the mid 90s [2]. However, the newly introduced automated indexing systems had to be evaluated to compare and improve the performance against benchmarks. The ongoing developments of the MTI then introduced machine learning components that have been tested across different document types, e.g., clinical health records that require different indexing approaches than merely assignment of MeSH Headings. The performance of the MTI on clinical health records has been evaluated in 2007 for the assignment of ICD-9 codes with promising results [3].
Solutions for the automated assignment of MeSH Headings have barely ever been evaluated, neither for their performance nor for their reproducibility. Conceptually, the evaluation of six different MeSH taggers showed that the k-nearest neighbour (k-NN) approach outperforms all other solutions [[33](#ref-CR33 "Trieschnigg, D., Pezik, P., Lee, V., de Jong, F., Kraaij, W., Rebholz-Schuhmann, D.: MeSH Up: effective MeSH text classification for improved document retrieval. Bioinformatics 25(11), 1412–1418 (2009). https://doi.org/10.1093/bioinformatics/btp249
")\]. Apart from the NLM’s critical response with regards to reproducibility, NLM still emphasizes “that current challenges in MeSH indexing include an increase of the scope of the task” \[[26](#ref-CR26 "Neveol, A., Mork, J., Aronson, A.: Comment on ‘MeSH-up: effective MeSH text classification for improved document retrieval’. Bioinformatics 25(20), 2770–2771 (2009).
https://doi.org/10.1093/bioinformatics/btp483
")\].The demands for such evaluation has motivated the NLM improving MTI as well as organizing large-scale evaluation challenges. Now MTI incorporates k-NN clustering showing a boost in the performance of the system [[15](#ref-CR15 "Jimeno-Yepes, A., Mork, J., Wilkowski, B., Demner-Fushman, D., Aronson, A.: MEDLINE mesh indexing: lessons learned from machine learning and future directions. In: ACM International Health Informatics Symposium, IHI 2012, Miami, FL, USA, pp. 737–742 (2012). https://doi.org/10.1145/2110363.2110450
")\], and in 2012, the BioASQ challenge was initiated (funding horizon of 5 years) leading to the evaluation of systems for large-scale biomedical indexing and question answering \[[34](#ref-CR34 "Tsatsaronis, G., et al.: BioASQ: a challenge on large-scale biomedical semantic indexing and question answering. In: Information Retrieval and Knowledge Discovery in Biomedical Text, Papers from the 2012 AAAI Fall Symposium, Arlington, Virginia, USA (2012).
http://www.aaai.org/ocs/index.php/FSS/FSS12/paper/view/5600
")\].2 Related Work
In 2013, the first BioASQ challenge was comprising two tasks, one on large-scale semantic indexing for the automated assignment of MeSH Headings to unlabeled Medline citations, the other one on question answering for scientific research questions in the biomedical domain [[27](#ref-CR27 "Partalas, I., Gaussier, É., Ngonga-Ngomo, A.: Results of the first BioASQ workshop. In: Proceedings of the First Workshop on Bio-Medical Semantic Indexing and Question at CLEF, Valencia, Spain (2013). http://ceur-ws.org/Vol-1094/bioasq2013_overview.pdf
")\]. In the first BioASQ challenge, 11 teams participated in Task A with 40 systems. In Task B, three different teams participated with 11 systems.In Task A, there were two baselines of Task A for large-scale semantic indexing, the first one was an unsupervised machine learning approach, the second one was based on NLM’s MTI. The evaluation was conducted using the metrics Micro F-measure (MiF) and Lowest Common Ancestor F-measure (LCA-F). The best-performing system, even outperforming the MTI baseline, called AUTH [[36](#ref-CR36 "Tsoumakas, G., Laliotis, M., Markantonatos, N., Vlahavas, I.: Large-scale semantic indexing of biomedical publications. In: Proceedings of the First Workshop on Bio-Medical Semantic Indexing and Question Answering at CLEF, Valencia, Spain (2013). http://ceur-ws.org/Vol-1094/bioasq2013_submission_6.pdf
")\], is based on a binary Support Vector Machine (SVM) predicting _N_ top labels for each article with a certain confidence score to rank the predicted labels.In Task B, two baselines were created as the top 50 and top 100 predictions of an ensemble system that combines predictions of factoid and list questions, yes/no questions, and summary questions. The evaluation metric was the Mean Average Precision (MAP). The Wishart [[10](#ref-CR10 "Cheng, D., Knox, C., Young, N., Stothard, P., Damaraju, S., Wishart, D.: PolySearch: a web-based text mining system for extracting relationships between human diseases, genes, mutations, drugs and metabolites. Nucleic Acids Res. 36(Web Server issue), 399–405 (2008). https://doi.org/10.1093/nar/gkn296
")\] system was able to outperform the two baselines. It uses the PolySearch[Footnote 3](#Fn3) tool for query expansion and the retrieval of candidate documents from which either entities or sentences are extracted as answers for the respective questions.The BioASQ challenge was then executed every year until today bringing about a variety of approaches in both tasks A and B [[5](#ref-CR5 "Balikas, G., Kosmopoulos, A., Krithara, A., Paliouras, G., Kakadiaris, I.: Results of the BioASQ tasks of the question answering lab at CLEF. In: Conference and Labs of the Evaluation forum, Toulouse, France (2015). http://ceur-ws.org/Vol-1391/inv-pap7-CR.pdf
"), [8](#ref-CR8 "Balikas, G., Partalas, I., Ngonga-Ngomo, A., Krithara, A., Paliouras, G.: Results of the BioASQ track of the question answering lab at CLEF. In: Working Notes for CLEF 2014 Conference, Sheffield, UK, pp. 1181–1193 (2014).
http://ceur-ws.org/Vol-1180/CLEF2014wn-QA-BalikasEt2014.pdf
"), [19](#ref-CR19 "Krithara, A., Nentidis, A., Paliouras, G., Kakadiaris, I.: Results of the 4th edition of BioASQ challenge. In: Proceedings of the Fourth BioASQ workshop, Berlin, Germany, pp. 1–7 (2016).
https://doi.org/10.18653/v1/W16-3101
"), [25](#ref-CR25 "Nentidis, A., Bougiatiotis, K., Krithara, A., Paliouras, G., Kakadiaris, I.: Results of the fifth edition of the BioASQ challenge. In: BioNLP 2017, Vancouver, Canada, pp. 48–57 (2017).
https://doi.org/10.18653/v1/W17-2306
")\]. In Task A, MeSHLabeler performed best the challenges 2014, 2015 and 2016 \[[21](#ref-CR21 "Liu, K., Peng, S., Wu, J., Zhai, C., Mamitsuka, H., Zhu, S.: MeSHLabeler: improving the accuracy of large-scale MeSH indexing by integrating diverse evidence. Bioinformatics 31(12), 339–347 (2015).
https://doi.org/10.1093/bioinformatics/btv237
")\] using an ensemble approach of k-NN, the MTI itself as well as further MeSH classification solutions.In recent years, term vector space representations have been introduced exceeding classical bag-of-words approaches, since they are able to capture the context of words in the text and to prioritize words in the vector representation according to given similarity scores [[24](#ref-CR24 "Mikolov, T., Sutskever, I., Chen, K., Corrad, G., Dean, J.: Distributed representations of words and phrases and their compositionality. CoRR abs/1310.4546 (2013). https://dblp.org/rec/bib/journals/corr/MikolovSCCD13
"), [30](#ref-CR30 "Pennington, J., Socher, R., Manning, C.: Glove: global vectors for word representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, EMNLP, Doha, Qatar, pp. 1532–1543 (2014).
https://dblp.org/rec/bib/conf/emnlp/PenningtonSM14
")\]. In addition, the word vectorization allows for better use of sentence and paragraph representations \[[20](#ref-CR20 "Le, Q., Mikolov, T.: Distributed representations of sentences and documents. arXiv preprint
arXiv:1405.4053
(2014).
https://arxiv.org/abs/1405.4053v2
")\] in the machine learning approaches, e.g., deep learning. The organizers of the BioASQ challenge also published a word2vec representation of PubMed articles \[[28](#ref-CR28 "Pavlopoulos, I., Kosmopoulos, A., Androutsopoulos, I.: Continuous space word vectors obtained by applying word2vec to abstracts of biomedical articles (2014).
http://bioasq.lip6.fr/info/BioASQword2vec/
")\] for participants to improve their systems.In 2017, the first deep learning based approach called DeepMeSH participated in the challenge of Task A and performed best [[29](#ref-CR29 "Peng, S., You, R., Wang, H., Zhai, C., Mamitsuka, H., Zhu, S.: DeepMeSH: deep semantic representation for improving large-scale MeSH indexing. Bioinformatics 32(12), 70–79 (2016). https://doi.org/10.1093/bioinformatics/btw294
")\]. In 2018, DeepMeSH outperformed others in 2 out of 3 batches while the third batch was won by a set of systems called “xgx” that is potentially associated to the AttentionMeSH system \[[16](#ref-CR16 "Jin, Q., Dhingra, B., Cohen, W., Lu, X.: AttentionMeSH: simple, effective and interpretable automatic MeSH indexer. In: Proceedings of the 6th BioASQ Workshop. A Challenge on Large-Scale Biomedical Semantic Indexing and Question Answering, pp. 47–56 (2018).
https://www.aclweb.org/anthology/W18-5306
")\]. This system uses end-to-end DeepLearning incorporating an attention layer to emphasize predictions towards commonly used MeSH labels.Further systems participated in the Task A employing named entity recognition with lexical features such as a dictionary, and using Paragraph Vectors also [[22](#ref-CR22 "Longwell, S.: Distributed representations for automating mesh indexing (2016). https://cs224d.stanford.edu/reports/Longwell.pdf
")\]. It has been shown that machine learning-based approaches based on k-NN and Paragraph Vectors can be used to boost the performance in the BioASQ challenge \[[17](#ref-CR17 "Kosmopoulos, A., Androutsopoulos, I., Paliouras, G.: Biomedical semantic indexing using dense word vectors in BioASQ (2015).
http://nlp.cs.aueb.gr/pubs/jbms_dense_vectors.pdf
")\]. This paper describes the participation of a system for Multi-Label Classification based on named entity recognition with lexical features that incorporate Label Ranking derived from Paragraph Vectors in order to achieve a conjoint system for Multi-Label Ranking.3 Methodology
Task A of the BioASQ challenge is a Multi-Label Classification task, which in addition can be subdivided into the two sub-tasks of Multi-Label Classification and Label Ranking [[35](#ref-CR35 "Tsoumakas, G., Katakis, I., Vlahavas, I.: Mining multi-label data. In: Data Mining and Knowledge Discovery Handbook, 2nd edn., pp. 667–685 (2010). https://doi.org/10.1007/978-0-387-09823-4_34
")\]. The resulting classification of multiple labels with an assigned confidence score for each label is a Multi-Label Ranking \[[9](#ref-CR9 "Brinker, K., Fürnkranz, J., Hüllermeier, E.: A unified model for multilabel classification and ranking. In: ECAI 2006, 17th European Conference on Artificial Intelligence, Riva del Garda, Italy, Proceedings, pp. 489–493 (2006).
https://dblp.org/rec/bib/conf/ecai/BrinkerFH06
")\].Initially, a subset of MeSH Headings is attributed to each document in the test set of Medline citations. Then, each MeSH Heading combined with its confidence score representing the probability for the MeSH Heading being correctly assigned to the respective Medline citation. The resulting set of MeSH Headings is filtered according to the minimum confidence score.
The two sub-tasks, i.e. Multi-Label Classification and Label Ranking for a Multi-Label Ranking, are given in the system architecture of this paper. The first component creates an initial set of MeSH Headings for each document in the test set for the Medline citations. Then, all MeSH Headings receive a confidence score to generate the scored MeSH Headings.
The first component for the task of Multi-Label Classification is described in Sect. 3.1 and the second component for the Label Ranking is described in Sect. 3.2. The combined system for the Multi-Label Ranking is described in Sect. 3.3.
3.1 Multi-label Classification
The Multi-Label Classification task is based on lexical features for the named entity recognition solution that has been developed within the Unstructured Information Management Architecture (UIMA)Footnote 4 [[11](#ref-CR11 "Ferrucci, D., Lally, A.: Accelerating corporate research in the development, application and deployment of human language technologies. In: Proceedings of the HLT-NAACL 2003 Workshop on Software Engineering and Architecture of Language Technology Systems (SEALTS), Morristown, NJ, USA, pp. 67–74 (2003). https://doi.org/10.3115/1119226.1119236
"),[12](#ref-CR12 "Ferrucci, D., Lally, A.: UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat. Lang. Eng. 10(3–4), 327–348 (2004).
https://doi.org/10.1017/S1351324904003523
"),[13](#ref-CR13 "Ferrucci, D., Lally, A., Verspoor, K., Nyberg, E.: Unstructured information management architecture (UIMA) version 1.0. OASIS Standard (2009).
https://docs.oasis-open.org/uima/v1.0/uima-v1.0.html
"),[14](#ref-CR14 "Götz, T., Suhre, O.: Design and implementation of the UIMA common analysis system. IBM Syst. J. 43(3), 476–489 (2004).
https://doi.org/10.1147/sj.433.0476
"), [31](#ref-CR31 "Schor, M.: An effective, Java-friendly interface for the unstructured management architecture (UIMA) common analysis system. Technical report IBM RC23176, IBM T. J. Watson Research Center (2004)")\]. In the framework, a reader for the BioASQ JSON format processes the document stream through the Common Analysis System (CAS) of the pipeline. Tokenization is conducted using an Offset Tokenizer that splits tokens at their whitespaces and punctuations. Stemming of the tokens is conducted using the Snowball Stemmer \[[1](#ref-CR1 "Agichtein, E., Gravano, L.: Snowball: extracting relations from large plain-text collections. In: Proceedings of the Fifth ACM Conference on Digital Libraries, pp. 85–94. ACM, New York (2000).
https://doi.org/10.1145/336597.336644
")\]. The stemmed tokens are analyzed using the analysis engine ConceptMapper \[[32](#ref-CR32 "Tanenblatt, M., Coden, A., Sominsky, I.: The ConceptMapper approach to named entity recognition. In: Proceedings of the International Conference on Language Resources and Evaluation, LREC, Valletta, Malta (2010).
http://www.lrec-conf.org/proceedings/lrec2010/summaries/448.html
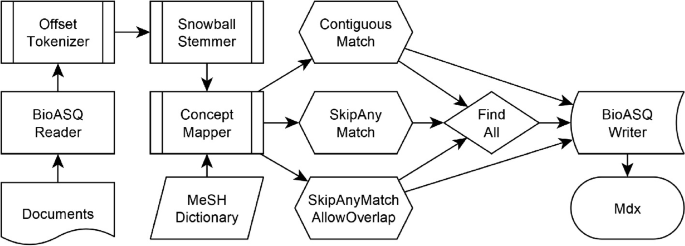
")\] that uses a dictionary to annotate matching synonyms in the text with offset information onto concept identifiers. In the last part of the UIMA-pipeline, the documents with their annotated MeSH Headings are written with a CAS-Writer into the BioASQ submission format. The implemented workflow is shown in Fig. [1](#Fig1).Fig. 1.
The UIMA-based workflow with different combinations of configurations for the ConceptMapper to produce the result set of MeSH Headings for Medline citations.
The lexical features for the ConceptMapper are provided as a dictionary that is created from the current MeSH (version 2019). In the dictionary, concepts are created for each MeSH Heading and synonyms are added from the MeSH Entry Terms for the MeSH Heading. Further synonyms are created with the Snowball Stemmer by stemming the concept name as well as each of the synonyms. The resulting dictionary for the ConceptMapper contains 29,351 different concepts with 251,463 synonyms.
The analysis engine ConceptMapper [[32](#ref-CR32 "Tanenblatt, M., Coden, A., Sominsky, I.: The ConceptMapper approach to named entity recognition. In: Proceedings of the International Conference on Language Resources and Evaluation, LREC, Valletta, Malta (2010). http://www.lrec-conf.org/proceedings/lrec2010/summaries/448.html
")\] provides various dictionary look-up solutions that can match against different sequences of tokens. Before applying a matching strategy, stop words and punctuation are removed. Then, one of the three lookup strategies are applied with a flag for allowing partial matches or allowing only complete matches. The different look-up solutions with the flag for finding also partial matches of synonyms will result in 5 different pattern matching configurations. For the BioASQ Task6a in 2018, each of the different dictionary look-up approaches are listed as separate system enumerated as SNOKE1 to SNOKE5\. For the BioASQ Task7a in 2019, all the results from the five different systems have been merged together into a union set.Fig. 2.
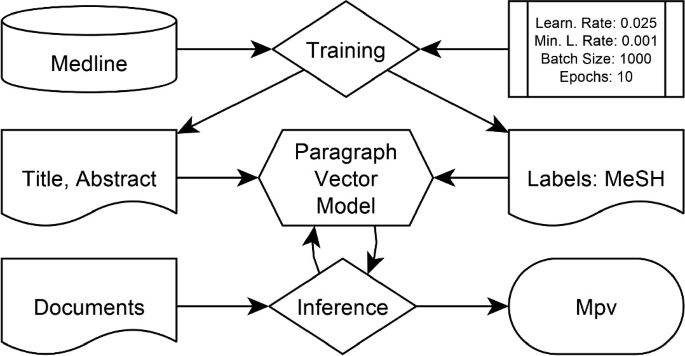
The Paragraph Vector model is trained on the BioASQ corpus with 225, 127 documents published from 2018 until January 2019 using the given MeSH Headings from the documents as training labels. Documents during the challenge were inferred using the trained model that resulted in a set \(M_{pv}\) of scored MeSH Headings for each document.
3.2 Label Ranking
Paragraph Vectors allow for capturing contextual information of words in text. The contextual information is trained by calculating the probability of certain words preceding or succeeding the contextual word. The resulting Paragraph Vector model enables the calculation of similarities of different texts according to their probability of occurring close to each other.
The task of Label Ranking is conducted by creating such a Paragraph Vector model to score all MeSH Headings for each document in the test set. Each MeSH Headings gets an assignment of a confidence score ranging from \(-1\) to 1. In order to provide such a system for assigning confidence scores, an unsupervised machine learning model is trained according to the algorithm described in [[20](#ref-CR20 "Le, Q., Mikolov, T.: Distributed representations of sentences and documents. arXiv preprint arXiv:1405.4053
(2014).
https://arxiv.org/abs/1405.4053v2
")\] (Fig. [2](#Fig2)).The Paragraph Vectors were trained using the BioASQ corpus with 225, 127 citations that have been published either in 2018 or 2019 in conjunction with their corresponding MeSH Headings of 25, 363 different labels in total. The Paragraph Vector model is PV-DBOW based on Skip-Grams (length 4) and have been trained using a configuration with 10 epochs, a learning rate of 0.025, a minimum learning rate of 0.001, and a batch size of 1000. The model is available onlineFootnote 5. The training as well as the predictions during the BioASQ challenge were implemented using the DeepLearning4j frameworkFootnote 6.
3.3 Multi-label Ranking
The task of Multi-Label Ranking is achieved by combining both systems for Multi-Label Classification and for Label Ranking. The first system uses five different vectors of MeSH Headings according to the pattern matching algorithm. For each document, a single set of MeSH Headings is created by taking the union set from the five different vectors.
Similarly, the Paragraph Vector model is used for assigning the confidence scores to each of the MeSH Headings. This results in a set of 25, 363 Headings for each document with a confidence score of \(-1\) to 1 assigned to each Heading. Then, both the union set as well as the confidence scores for the MeSH Headings are joined by filtering the union set for only the top-k scored terms. K was chosen for 500 and for 1, 0000 resulting in two different BioASQ Task7a submissions. The algorithm for creating the sets \(M_{top500}\) and \(M_{top1000}\) for each document is shown in Algorithm 1.

4 Results
In BioASQ TaskA, the systems have been challenged to outperform the MTI for the annotation of Medline citations with MeSH Headings. In the challenge, there have been three test batches leading to 5 runs for each batch. For each run, a test set of Medline citations has been published that have not yet been annotated with MeSH Headings by human curators. The evaluation of the participating systems for each of the runs in every batch is an automated process implemented within the BioASQ infrastructure [6, 7].
The evaluation infrastructure computes the results with two different classes of measurements, flat and hierarchical. The comparison of the performance of the participating systems are assessed with one flat and one hierarchical measure: the Lowest Common Ancestor F1-measure LCA.F [[18](#ref-CR18 "Kosmopoulos, A., Partalas, I., Gaussier, É., Paliouras, G., Androutsopoulos, I.: Evaluation measures for hierarchical classification: a unified view and novel approaches. CoRR abs/1306.6802 (2013). https://dblp.org/rec/bib/journals/corr/KosmopoulosPGPA13
")\] and the Label-Based Micro F1-measure MiF. Besides the two main evaluation F1-measures, there is also the Example-Based F1-Measure, Accuracy, Label-Based Macro F1-Measure, and Hierarchical F1-Measure. For each F1-Measure, the respective precision and recall measures are calculated.The system for the Multi-Label Classification participated in the Task6a in 2018 and is explained in Sect. 4.1. The conjoint system that uses the initial Multi-Label Classification for Label-Ranking in order to produce a Multi-Label Ranking participated in the Task7a in 2019 and is explained in Sect. 4.2.
Fig. 3.
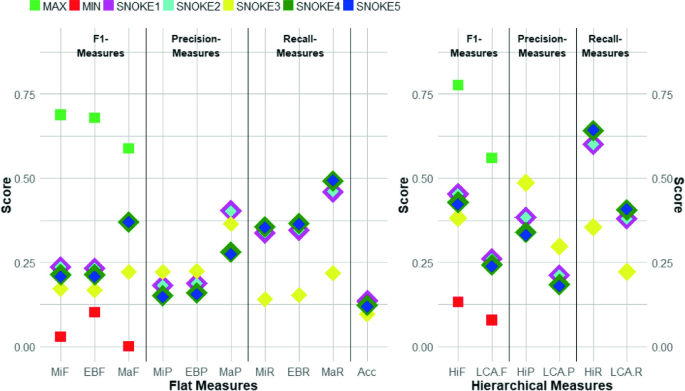
The 10 flat and the 6 hierarchical measures are sorted according to the F1 measure, Precision, and Recall for each of the different configuration setups in Task6a. The maximum score achieved by the best participating system is given as green squares, the minimum score achieved by the worst participating system as red squares. (Color figure online)
4.1 System for Multi-label Classification in Task6a
In the 2018 BioASQ Task6a, the maximum MiF score of 0.6880 was achieved by the system xgx and the maximum LCA-F score of 0.5596 was achieved by the system xgx0. The maximum label-based micro-recall (MiR) was 0.6751 and the maximum label-based micro-precision (MiP) was 0.8110. The highest lowest common ancestor recall (LCA-R) was 0.5563 and the highest lowest common ancestor precision (LCA-P) was 0.6212. For all participating systems, the tendency was towards having a higher precision than having a higher recall.
Contrastingly, the submissions for the Multi-Label Classification system with SNOKE1 to SNOKE5 reached higher recall measures than precision measures. The maximum MiF score of 0.236 was achieved by both the submissions for SNOKE1 and SNOKE2. The maximum LCA-F score of 0.261 was achieved by the submission for SNOKE1. The highest MiR was 0.356 while the MiP was 0.221. A similar picture was shown for the highest LCA-R having a 0.408 while the highest LCA-P was 0.298. In Fig. 3, the 10 flat and the 6 hierarchical measurements for SNOKE1 to SNOKE5 are visualized.
Fig. 4.
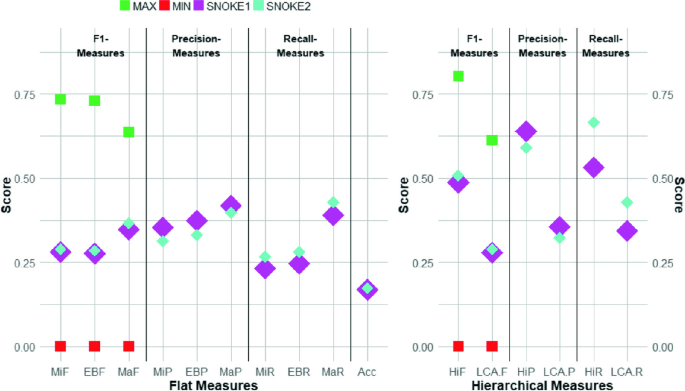
The 10 flat and 6 hierarchical measures arranged according to their F1-Measure, Precision, and Recall for each configuration setup in Task7a. The maximum score achieved by the best participating system as green squares, the minimum score achieved by the worst participating system as red squares. (Color figure online)
4.2 System for Multi-label Ranking in Task7a
In the 2019 BioASQ Task7a, both the maximum MiF score of 0.733 and the maximum LCA-F score of 0.612 was achieved by the system DeepMeSH5. The maximum (MiR) was 0.707 and the maximum MiP was 0.791. The highest LCA-R was 0.6 and the highest LCA-P was 0.663. Similar to the Task6a in 2018, the tendency was again more towards having a higher precision than having a higher recall.
In comparison to the 2018 participation, the submissions for the Multi-Label Ranking system SNOKE1 and SNOKE2 reached higher precision measures than for the recall measures for the label-based micro-measures. The maximum MiF score was 0.288 with a maximum MiP of 0.354 and a maximum MiR of 0.267. For the measures of the lowest common ancestor, the recall was higher than the precision. The maximum \(LCA-F\) score was 0.288 with a maximum LCA-P of 0.356 and a maximum LCA-R with 0.428. In Fig. 4, the 10 flat and the 6 hierarchical measurements for the submissions of SNOKE1 and SNOKE2 are visualized.
5 Conclusion
This paper describes the participation of two different systems and their combined solution in the Task6a and the Task7a of the BioASQ challenge. The first system that participated in Task6a is a Multi-Label Classification system that incorporates lexical features from MeSH. The system was extended for the participation in the Task7a for the functionality of introducing Label Ranking for the assignment of confidence scores to MeSH Headings resulting in a conjoint system for Multi-Label Ranking.
The first system for Multi-Label Classification participated in the Task6a of the BioASQ challenge. The results indicate that the recall for the system are higher than the precision although the general tendency of the other participating systems is the opposite. Nevertheless, the label-based macro F1-measure shows better performance than the label-based micro F1-measure.
The second system that incorporates both Multi-Label Classification and Label-Ranking for Multi-Label Ranking participated in the Task7a of the BioASQ challenge. All performance measures were improved in comparison to the first system that participated in the Task6a of the BioASQ challenge. Generally, the precision has been boosted in comparison to the earlier participation.
6 Discussion
In the BioASQ challenge, systems are supposed to outperform the baseline of the MTI for the assignment of MeSH Headings to Medline citations. Two different participations in the BioASQ challenge are described in this paper, one for Task6a and one for Task7a. The initially developed system that incorporates lexical features from the MeSH thesaurus is extended to a ranking of MeSH Headings according to the confidence values.
The participation of the first system in Task6a generally shows higher recall than precision performance. The extended system that exploits the ranking of MeSH Headings was able to increase both, precision and recall, resulting in better F1-measures overall. However, the initially assigned MeSH Headings based on lexical features still have a low overlap in comparison to the assigned MeSH Headings by the human curators.
References
- Agichtein, E., Gravano, L.: Snowball: extracting relations from large plain-text collections. In: Proceedings of the Fifth ACM Conference on Digital Libraries, pp. 85–94. ACM, New York (2000). https://doi.org/10.1145/336597.336644
- Aronson, A., et al.: The NLM indexing initiative. In: AMIA 2000, American Medical Informatics Association Annual Symposium, Los Angeles, CA, USA, 4–8 November 2000 (2000)
Google Scholar - Aronson, A., et al.: From indexing the biomedical literature to coding clinical text: experience with MTI and machine learning approaches. In: Biological, Translational and Clinical Language Processing, BioNLP@ACL Prague, Czech Republic, pp. 105–112 (2007)
Google Scholar - Aronson, A., Mork, J., Gay, C., Humphrey, S., Rogers, W.: The NLM indexing initiative’s medical text indexer. Stud. Health Technol. Inform. 107, 268–272 (2004)
Google Scholar - Balikas, G., Kosmopoulos, A., Krithara, A., Paliouras, G., Kakadiaris, I.: Results of the BioASQ tasks of the question answering lab at CLEF. In: Conference and Labs of the Evaluation forum, Toulouse, France (2015). http://ceur-ws.org/Vol-1391/inv-pap7-CR.pdf
- Balikas, G., Partalas, I., Baskiotis, N., Artieres, T., Gausier, E., Gallinari, P.: Evaluation infrastructure software for the challenges 2nd version. Technical report D4.7 (2014)
Google Scholar - Balikas, G., Partalas, I., Baskiotis, N., Artieres, T., Gaussier, E., Gallinari, P.: Evaluation infrastructure. Technical report (2013)
Google Scholar - Balikas, G., Partalas, I., Ngonga-Ngomo, A., Krithara, A., Paliouras, G.: Results of the BioASQ track of the question answering lab at CLEF. In: Working Notes for CLEF 2014 Conference, Sheffield, UK, pp. 1181–1193 (2014). http://ceur-ws.org/Vol-1180/CLEF2014wn-QA-BalikasEt2014.pdf
- Brinker, K., Fürnkranz, J., Hüllermeier, E.: A unified model for multilabel classification and ranking. In: ECAI 2006, 17th European Conference on Artificial Intelligence, Riva del Garda, Italy, Proceedings, pp. 489–493 (2006). https://dblp.org/rec/bib/conf/ecai/BrinkerFH06
- Cheng, D., Knox, C., Young, N., Stothard, P., Damaraju, S., Wishart, D.: PolySearch: a web-based text mining system for extracting relationships between human diseases, genes, mutations, drugs and metabolites. Nucleic Acids Res. 36(Web Server issue), 399–405 (2008). https://doi.org/10.1093/nar/gkn296
Article Google Scholar - Ferrucci, D., Lally, A.: Accelerating corporate research in the development, application and deployment of human language technologies. In: Proceedings of the HLT-NAACL 2003 Workshop on Software Engineering and Architecture of Language Technology Systems (SEALTS), Morristown, NJ, USA, pp. 67–74 (2003). https://doi.org/10.3115/1119226.1119236
- Ferrucci, D., Lally, A.: UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat. Lang. Eng. 10(3–4), 327–348 (2004). https://doi.org/10.1017/S1351324904003523
Article Google Scholar - Ferrucci, D., Lally, A., Verspoor, K., Nyberg, E.: Unstructured information management architecture (UIMA) version 1.0. OASIS Standard (2009). https://docs.oasis-open.org/uima/v1.0/uima-v1.0.html
- Götz, T., Suhre, O.: Design and implementation of the UIMA common analysis system. IBM Syst. J. 43(3), 476–489 (2004). https://doi.org/10.1147/sj.433.0476
Article Google Scholar - Jimeno-Yepes, A., Mork, J., Wilkowski, B., Demner-Fushman, D., Aronson, A.: MEDLINE mesh indexing: lessons learned from machine learning and future directions. In: ACM International Health Informatics Symposium, IHI 2012, Miami, FL, USA, pp. 737–742 (2012). https://doi.org/10.1145/2110363.2110450
- Jin, Q., Dhingra, B., Cohen, W., Lu, X.: AttentionMeSH: simple, effective and interpretable automatic MeSH indexer. In: Proceedings of the 6th BioASQ Workshop. A Challenge on Large-Scale Biomedical Semantic Indexing and Question Answering, pp. 47–56 (2018). https://www.aclweb.org/anthology/W18-5306
- Kosmopoulos, A., Androutsopoulos, I., Paliouras, G.: Biomedical semantic indexing using dense word vectors in BioASQ (2015). http://nlp.cs.aueb.gr/pubs/jbms_dense_vectors.pdf
- Kosmopoulos, A., Partalas, I., Gaussier, É., Paliouras, G., Androutsopoulos, I.: Evaluation measures for hierarchical classification: a unified view and novel approaches. CoRR abs/1306.6802 (2013). https://dblp.org/rec/bib/journals/corr/KosmopoulosPGPA13
- Krithara, A., Nentidis, A., Paliouras, G., Kakadiaris, I.: Results of the 4th edition of BioASQ challenge. In: Proceedings of the Fourth BioASQ workshop, Berlin, Germany, pp. 1–7 (2016). https://doi.org/10.18653/v1/W16-3101
- Le, Q., Mikolov, T.: Distributed representations of sentences and documents. arXiv preprint arXiv:1405.4053 (2014). https://arxiv.org/abs/1405.4053v2
- Liu, K., Peng, S., Wu, J., Zhai, C., Mamitsuka, H., Zhu, S.: MeSHLabeler: improving the accuracy of large-scale MeSH indexing by integrating diverse evidence. Bioinformatics 31(12), 339–347 (2015). https://doi.org/10.1093/bioinformatics/btv237
Article Google Scholar - Longwell, S.: Distributed representations for automating mesh indexing (2016). https://cs224d.stanford.edu/reports/Longwell.pdf
- Lu, Z.: PubMed and beyond: a survey of web tools for searching biomedical literature. Database (Oxford), p. baq036 (2011). https://doi.org/10.1093/database/baq036
- Mikolov, T., Sutskever, I., Chen, K., Corrad, G., Dean, J.: Distributed representations of words and phrases and their compositionality. CoRR abs/1310.4546 (2013). https://dblp.org/rec/bib/journals/corr/MikolovSCCD13
- Nentidis, A., Bougiatiotis, K., Krithara, A., Paliouras, G., Kakadiaris, I.: Results of the fifth edition of the BioASQ challenge. In: BioNLP 2017, Vancouver, Canada, pp. 48–57 (2017). https://doi.org/10.18653/v1/W17-2306
- Neveol, A., Mork, J., Aronson, A.: Comment on ‘MeSH-up: effective MeSH text classification for improved document retrieval’. Bioinformatics 25(20), 2770–2771 (2009). https://doi.org/10.1093/bioinformatics/btp483
Article Google Scholar - Partalas, I., Gaussier, É., Ngonga-Ngomo, A.: Results of the first BioASQ workshop. In: Proceedings of the First Workshop on Bio-Medical Semantic Indexing and Question at CLEF, Valencia, Spain (2013). http://ceur-ws.org/Vol-1094/bioasq2013_overview.pdf
- Pavlopoulos, I., Kosmopoulos, A., Androutsopoulos, I.: Continuous space word vectors obtained by applying word2vec to abstracts of biomedical articles (2014). http://bioasq.lip6.fr/info/BioASQword2vec/
- Peng, S., You, R., Wang, H., Zhai, C., Mamitsuka, H., Zhu, S.: DeepMeSH: deep semantic representation for improving large-scale MeSH indexing. Bioinformatics 32(12), 70–79 (2016). https://doi.org/10.1093/bioinformatics/btw294
Article Google Scholar - Pennington, J., Socher, R., Manning, C.: Glove: global vectors for word representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, EMNLP, Doha, Qatar, pp. 1532–1543 (2014). https://dblp.org/rec/bib/conf/emnlp/PenningtonSM14
- Schor, M.: An effective, Java-friendly interface for the unstructured management architecture (UIMA) common analysis system. Technical report IBM RC23176, IBM T. J. Watson Research Center (2004)
Google Scholar - Tanenblatt, M., Coden, A., Sominsky, I.: The ConceptMapper approach to named entity recognition. In: Proceedings of the International Conference on Language Resources and Evaluation, LREC, Valletta, Malta (2010). http://www.lrec-conf.org/proceedings/lrec2010/summaries/448.html
- Trieschnigg, D., Pezik, P., Lee, V., de Jong, F., Kraaij, W., Rebholz-Schuhmann, D.: MeSH Up: effective MeSH text classification for improved document retrieval. Bioinformatics 25(11), 1412–1418 (2009). https://doi.org/10.1093/bioinformatics/btp249
Article Google Scholar - Tsatsaronis, G., et al.: BioASQ: a challenge on large-scale biomedical semantic indexing and question answering. In: Information Retrieval and Knowledge Discovery in Biomedical Text, Papers from the 2012 AAAI Fall Symposium, Arlington, Virginia, USA (2012). http://www.aaai.org/ocs/index.php/FSS/FSS12/paper/view/5600
- Tsoumakas, G., Katakis, I., Vlahavas, I.: Mining multi-label data. In: Data Mining and Knowledge Discovery Handbook, 2nd edn., pp. 667–685 (2010). https://doi.org/10.1007/978-0-387-09823-4_34
- Tsoumakas, G., Laliotis, M., Markantonatos, N., Vlahavas, I.: Large-scale semantic indexing of biomedical publications. In: Proceedings of the First Workshop on Bio-Medical Semantic Indexing and Question Answering at CLEF, Valencia, Spain (2013). http://ceur-ws.org/Vol-1094/bioasq2013_submission_6.pdf
Author information
Authors and Affiliations
- ZB MED - Information Centre for Life Sciences, Gleueler Str. 60, 50931, Cologne, Germany
Bernd Müller & Dietrich Rebholz-Schuhmann
Authors
- Bernd Müller
You can also search for this author inPubMed Google Scholar - Dietrich Rebholz-Schuhmann
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toBernd Müller .
Editor information
Editors and Affiliations
- Institut National des Sciences Appliquées, Rennes, France
Peggy Cellier - Maastricht University, Maastricht, The Netherlands
Kurt Driessens
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Müller, B., Rebholz-Schuhmann, D. (2020). Selected Approaches Ranking Contextual Term for the BioASQ Multi-label Classification (Task6a and 7a). In: Cellier, P., Driessens, K. (eds) Machine Learning and Knowledge Discovery in Databases. ECML PKDD 2019. Communications in Computer and Information Science, vol 1168. Springer, Cham. https://doi.org/10.1007/978-3-030-43887-6\_52
Download citation
- .RIS
- .ENW
- .BIB
- DOI: https://doi.org/10.1007/978-3-030-43887-6\_52
- Published: 28 March 2020
- Publisher Name: Springer, Cham
- Print ISBN: 978-3-030-43886-9
- Online ISBN: 978-3-030-43887-6
- eBook Packages: Computer ScienceComputer Science (R0)