Impact of digestive and oropharyngeal decontamination on the intestinal microbiota in ICU patients (original) (raw)
Abstract
Purpose
Selective digestive microbial decontamination (SDD) is hypothesized to benefit patients in intensive care (ICU) by suppressing Gram-negative potential pathogens from the colon without affecting the anaerobic intestinal microbiota. The purpose of this study was to provide more insight to the effects of digestive tract and oropharyngeal decontamination on the intestinal microbiota by means of a prospective clinical trial in which faecal samples were collected from ICU patients for intestinal microbiota analysis.
Methods
The faecal samples were collected from ICU patients enrolled in a multicentre trial to study the outcome of SDD and selective oral decontamination (SOD) in comparison with standard care (SC). Fluorescent in situ hybridization (FISH) was used to analyze the faecal microbiota. The numbers of bacteria from different bacterial groups were compared between the three regimens.
Results
The total counts of bacteria per gram faeces did not differ between regimens. The F. prausnitzii group of bacteria, representing an important group among intestinal microbiota, was significantly reduced in the SDD regimen compared to the SC and SOD. The Enterobacteriaceae were significantly suppressed during SDD compared to both SOD and SC; enterococci increased in SDD compared to both other regimens.
Conclusions
The composition of the intestinal microbiota is importantly affected by SDD. The F. prausnitzii group was significantly suppressed during SDD. This group of microbiota is a predominant producer of butyrate, the main energy source for colonocytes. Reduction of this microbiota is an important trade-off while reducing gram-negative bacteria by SDD.
Similar content being viewed by others
Introduction
Selective (microbial) decontamination of the digestive tract (SDD), developed using immuno-compromised animal models [1, 2], was first clinically tested in severely immuno-compromised hemato-oncological patients and later applied to patients admitted to intensive care units (ICU) [3]. The concept of SDD is to selectively suppress potential pathogens, mostly Gram-negative bacteria, without disturbing the anaerobic intestinal microbiota. Oral, non-absorbable antibiotics were combined with prophylactic systemic antimicrobial treatment (3rd generation cephalosporins) during the first four days to eradicate potential pathogens. In order to protect the anaerobic microbiota, the use of antibiotics with anti-anaerobic activity was discouraged.
Earlier studies were flawed by design [3] or lacked statistical power to detect a survival advantage. Meta-analyses showed a significant survival advantage [4, 5] one did not [6]. Also single centre randomized trials evaluating SDD showed reduction in mortality [7, 8].
A consistent finding across studies evaluating SDD in the ICU has been a reduction of the number of episodes of nosocomial infections—especially, of respiratory tract. As the prevention of ventilator-associated pneumonia (VAP) might play a dominant role in the mortality reduction of SDD, the oropharyngeal component of SDD, referred to as Selective Oropharyngeal Decontamination (SOD), has also been analysed in clinical trials [9]. Although a significant reduction in VAP was shown, historically no overall reduction in mortality by SOD [10] or non-selective oral decontamination [11] was observed.
A large multi-centre clinical trial of SOD and SDD was recently reported. In this trial both SDD and SOD showed a similar survival benefit compared to standard care (SC) [12]. As an adjunct to this study we investigated the impact of SOD and SDD regimes on the intestinal microbiota compared to SC.
The intestinal microbiota is a complex ecosystem which comprises more than 1011 bacteria per gram of faeces and more than 400 different species [13]. Some of the intestinal microbiota are beneficial and ways to promote their growth have been investigated [14]. With the use of SDD, the intestinal microbiota is believed to protect the human host by preventing increased colonisation with potential pathogens [15], mostly anaerobic bacteria that are difficult to isolate and identify by classical culture techniques. Quantification of the anaerobic microbiota based on culture methods is unreliable: selective media introduce bias; and many genera cannot be cultured in vitro. Molecular methods, such as fluorescent in situ hybridisation (FISH), yield absolute numbers of micro-organisms [16, 17] instead of colony-forming units, which is the quantitative read-out of culture.
In the present study we evaluate, for the first time with molecular methods, the impact of SDD and SOD on the intestinal microbiota compared to SC in subjects admitted to ICU. We tested the hypothesis that SDD (or SOD) may execute its claimed beneficial effects by leaving the anaerobic intestinal microbiota unaffected.
Patients and methods
All patients consecutively admitted to the Medical ICU in our hospital, and that were evaluated within the framework of the Dutch multi-centre SDD-SOD study [12], were eligible. In this SDD-SOD study, participating centres followed three different regimens of treatment in their ICUs in a non-blinded random sequence:
- SC: no prophylactic antimicrobials, no restrictions in the antibiotics used
- SOD: prophylactic topical oropharyngeal antimicrobials, no restrictions in the antibiotics used.
- SDD: prophylactic topical oropharyngeal and gastro-intestinal antimicrobials, with the addition of intravenous cefotaxime during the first four days. Selective use of antimicrobials was encouraged to avoid interference with the intestinal microbiota, in accordance with the concept of colonisation resistance.
The sequence of these courses allocated to our hospital was SC–SOD–SDD. For details of the trial and the antibiotic regimens used, we refer to the original article [12]. All included patients were scored for their severity of disease with the APACHE II classification system [18]. Other parameters that were recorded are demographics (age, gender etc.), concomitant diseases and the use of all antimicrobial products including the antimicrobials used for the prophylactic protocols (SOD and SDD).
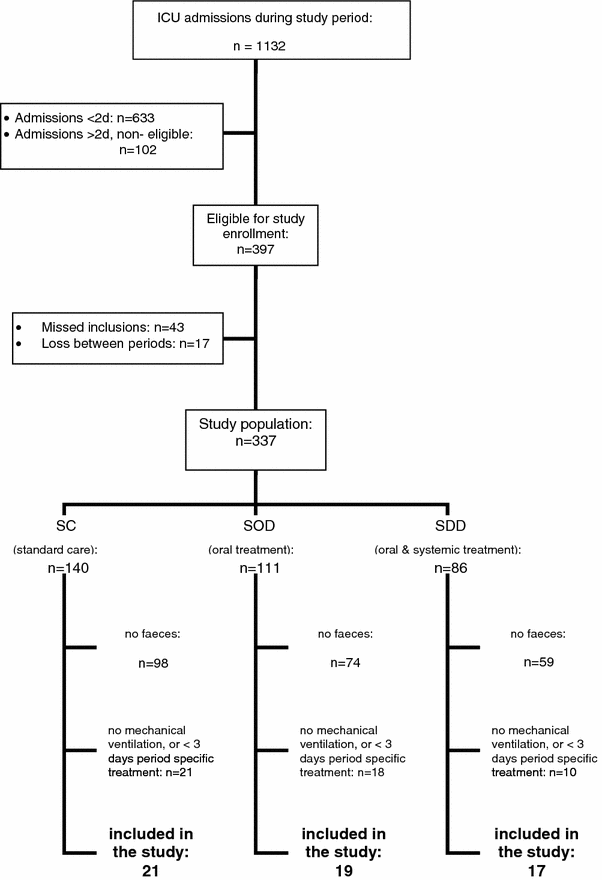
During all three consecutive trial periods, faecal samples were collected from the patients that produced faeces spontaneously or with the use of laxatives as on clinical indication alone. For every patient, only the first sample faecal that was passed after at least three days of period-specific treatment by patients on mechanical ventilation was used in order to avoid skewing of data (see Fig. 1).
Fig. 1
Flow chart of patient selection
For the multi-centre SOD-SDD study, a waiver from informed consent was provided by local and National Ethics Review Boards, as not patients but rather different standards of treatment protocols were compared in a randomized fashion. Samples and data were analyzed anonymously.
Materials and methods
All fresh faecal samples were stored in a refrigerator and processed for analysis within 24 h after collection. The processing procedure was as follows: after homogenization, from each faecal sample 1.0 g (wet weight) was taken and diluted in PBS (8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4·2H2O, 0.24 g/L KH2PO4, pH 7.4) 1:4 or 1:9 depending on faecal consistency. The faecal consistency was scaled on the Bristol Stool chart, where consistencies of stool type 1–6 were diluted 1:9, type 7 was diluted 1:4. These dilutions were processed for storage as described previously [19].
For quantification of the bacteria in faecal samples, multiple slides with 1 cm2 wells were prepared for cell counting and hybridized as described previously [19]. Hybridization was performed using the 16S rRNA-targeted probes listed in table 1.
Table 1 Probes used for the detection of the intestinal microbiota
The probes were manufactured by Eurogentec (Seraing, Belgium). Together these probes detect approximately 90% of the expected hybridisable total amount of bacteria (Eub338) [17]. Additional to the probe set for detection of the normal intestinal microbiota, three probes were used to enumerate groups of potential pathogens. These are the EC1531 [20] probe for the Enterobacteriaceae and the Enfl84 and Enfm93 [21] to detect enterococci.
The fluorescent cells in the samples were counted using an automated microscope system [22]. The detection limit used with this method was 106 cells/g of faeces.
After analysis of the FISH results, minimal inhibitory concentrations (MIC) to the SDD/SOD antibiotics of a type strain of F. prausnitzii were performed. The A2-165 strain of F. prausnitzii was cultured in an anaerobic cabinet on YCFA agar-plates. YCFA medium consisted of (per 100 ml): 1 g casitone, 0.25 g yeast extract, 0.4 g NaHCO3, 0.1 g cysteine, 0.045 g K2HPO4, 0.045 g KH2PO4, 0.09 g NaCl, 0.009 g MgSO4.7H2O, 0.009 g CaCl2, 0.1 mg resazurin, 1 mg haemin, 1 μg biotin, 1 μg cobalamin, 3 μg _p_-aminobenzoic acid, 5 μg folic acid and 15 μg pyridoxamine. Final concentrations of short-chain fatty acids (SCFA) in the medium were 33 mM acetate, 9 mM propionate and 1 mM each of isobutyrate, isovalerate and valerate.
The MIC tests were performed using E-test® -strips containing tobramycin, colistin and cefotaxime, according to the instructions of the manufacturer (AB BIODISK, Solna, Sweden).
Statistics
The results were compared statistically between the three patient groups using SPSS® 16 statistical analysis software. For the continuous variables age and APACHE-scores, ANOVA analyses were performed, for nominal variables such as antibiotics use, a Chi-square cross tabulation with a Fisher’s Exact test was used (see Table 2 for details).
Table 2 Characteristics of the patient population
The analysis of the numbers of bacteria was done with an ANOVA analysis after log-transformation to obtain a normal distribution. The normal distribution was checked with the Kolmogorov-Smirnoff test and by evaluation of P–P plots.
The numbers of enterococci however were still not normally distributed after log-transformation, therefore for these bacterial groups non-parametric tests (MWU) were performed.
Results
Patients: Figure 1 shows the flowchart of the patient sample selection. Faecal samples were collected for analysis from a total of 21 patients in the SC regimen, 19 patients in the SOD regimen and 17 patients in the SDD regimen.
The age distribution of patients did not differ significantly between the three study episodes; in the SC group, the mean age was 59.8, in the SOD group 63.7 and in the SDD group 56.7.
APACHE-II scores were also similar between groups: 14.3, 15.4 and 16.2 for the SC, SOD and SDD groups, respectively. The APACHE predicted (adjusted) death rate was higher in both the SOD and SDD regimen groups. These differences were not statistically significant. Furthermore enteral tube feeding and gastric retention did not differ statistically significantly between the three regimen groups, see table 2 for details.
Except for the use of cephalosporins (which are a part of the SDD regimen) the use of antibiotics did not differ significantly between the regimens. Because no statistical difference in the characteristics of the population was found, no further multivariate analysis was performed.
No significant impact of SDD was observed on the total number of bacteria of the colonic microbiota (Table 3). There is, however, a significant difference in the composition of the microbiota of the different regimens. Some differences are seen in the bacterial groups between the analysed regimens, but the numbers of the F. prausnitzii group in the SDD-regimen are significantly lower compared to both Standard Care and the SOD regimen. The Eubacterium rectale group shows lower numbers of bacteria in the SDD regimen compared to the SC regimen. A subgroup of _E. rectale_—the Roseburia intestinalis group—also shows lower numbers in the SDD regimen.
Table 3 Numbers and statistical analysis of the main intestinal microbiota groups
SDD had a significant impact on potential Gram-negative pathogen counts in the faecal microbiota to which it is targeted. Enterobacteriaceae in the SDD regimen were significantly reduced in numbers compared to both the SC-regimen and the SOD-regimen.
The gram-positive potential pathogens such as the enterococci increased significantly with the use of SDD, in comparison with both other regimens (Table 4). A significant rise in Enterococcus faecalis was also seen in the SOD regimen compared to the SC regimen. However, the numbers of E. faecalis in the SOD regimen were still significantly lower than when compared to the numbers of E. faecalis in the SDD regimen.
Table 4 Numbers and statistical analysis of enterococci per gram faeces
The MIC values of F. prausnitzii A2-165 for the SDD/SOD antibiotics were 4 μg/ml for tobramycin, >32 μg/ml for cefotaxime and >256 μg/ml for colistin.
Discussion
The major finding in this study was that the composition of the intestinal microbiota, as evidenced by stool analysis, was affected by the use of SDD: the F. prausnitzii group was significantly reduced in numbers in stools of subjects receiving SDD compared to subjects in both other regimens. This finding contrasts with the hypothesis of SDD that anaerobic microbiota would remain unaffected [15].
The group of bacteria detected with the Fprau-probe is one of the predominant bacterial groups in healthy volunteers, representing 10–15% of the intestinal microbiota on average [17, 23]. The F. prausnitzii group therefore plays an important role in maintaining the colonization resistance, normally protecting the human host from infections. F. prausnitzii has also been found to have anti-inflammatory effects which may play a dominant role in the development of Crohn's disease [24].
Furthermore, the F. prausnitzii group is considered to provide special health benefit for the human host because of its main fermentation product, butyrate [23, 25]. Butyrate is a short chain fatty acid that is the primary source of energy for the colonocytes [26]. In addition, butyrate promotes the growth of colonocytes, preventing mucosal atrophy. It also appears to lower the risk of malignant transformation of colonocytes in animal models [27]. The optimal concentration of butyrate is not known in vivo, but in vitro cell cultures show a growth arrest at concentrations below 10 mM [26]. In healthy volunteers, butyrate concentrations were also reduced during tube feeding [28], and in a blinded re-analysis of these faecal samples, the reduction of F. prausnitzii showed strong correlation with the reduction of butyrate measured in these faecal samples (manuscript submitted).
The _F. prausnitzii_-group of bacteria is difficult to culture in vitro and therefore could not be detected when SDD was first developed. Only culture-based methods were used to asses whether the intestinal microbiota remained intact in the early days of SDD. Therefore, this unintended effect could not have been foreseen when the regimen was first used. The MIC analysis of the SDD/SOD antibiotics shows that F. prausnitzii is susceptible only to tobramycin at 4 μg/ml. This concentration of tobramycin is easily reached with the intestinal decontamination regimen of SDD. The reduction of F. prausnitzii is therefore most likely caused by the intestinal administration of tobramycin in the SDD-regimen group.
We show that the loss of F. prausnitzii is not compensated for by an increase of other important butyrate-producing bacteria. Roseburia spp., another important group of butyrate producing bacteria, are also present in significantly lower numbers during the SDD regimen compared to the SC regimen. Similar to F. prausnitzii, reductions in numbers of bacteria from the Roseburia spp. correlate to a reduction in the amount of butyrate which is produced [29]. Therefore, we postulate that butyrate production may be impaired in SDD patients due to the loss of these predominant butyrate-producing bacterial groups.
Clear evidence that the SDD regimen was given according to protocol is the fact that a significant reduction in the Enterobacteriaceae was found in the SDD regimen compared to both other regimens. The numbers of Enterobacteriaceae in the SC and SOD regimens were markedly higher than in healthy individuals [17].
Traditionally, cefotaxime been a component of the SDD regimen [8, 12, 30]. Although cefotaxime has been shown to have a moderate suppressive effect on Enterobacteriaceae [31, 32], we have reasons to believe that the effect of cefotaxime is negligible compared to the effect of the large amounts of non-absorbable antibiotics targeted specifically to Enterobacteriaceae.
Cefotaxime elimination is almost entirely by renal excretion; only 5% of elimination is by biliary excretion, and the amount of cefotaxime reaching the colon is exceedingly low compared to the amounts of non-resorbable tobramycin and colistin, both active against Enterobacteriaceae. Early studies haven shown that cefotaxime is almost entirely inactivated by faecal enzymes [33]. This is not the case with colistin and tobramycin, which makes it most likely that the effect seen is caused by these antibiotics instead. Furthermore the earlier studies of the effects of cefotaxime on the intestinal microbiota are based upon culturing methods with selective plates. Especially when antibiotics are used, these methods can lead to underestimation of the actual numbers of bacteria.
The second important finding is that SOD left the faecal microbiota relatively unaffected compared to stools from patients enrolled during the SC period of the study. As shown in the multi-center clinical trial [12], there is no significant difference in mortality between SDD and SOD, despite the fact that colonisation and infections with Enterobacteriaceae are significantly reduced in SDD compared to SOD. This reduction of Enterobacteriaceae is confirmed in our study. We speculate that the lack of further mortality reduction is partly explained by the negative effect of SDD on beneficial bacteria of the colonic microbiota. How the loss of these beneficial bacteria, which provide an important source of nutrition for colonocytes, translates into clinically significant effects in general, and in critically ill patients in particular, is presently unknown. The role of microbiota and the production and uptake of butyrate in particular to maintain colonic integrity in a range of conditions, including critical care settings with sepsis, should perhaps be studied in animal models.
The Bifidobacteria showed a mild increase in the SDD regimen that did not reach statistical significance. These Gram-positive bacteria are not susceptible to the antibiotics used in SDD.
A third consideration with the use of SDD is the increase of enterococci in the faeces [34]. Although the numbers of E. faecalis are also significantly higher in the SOD regimen compared to the SC regimen, the numbers of enterococci in the SDD regimen are an order of magnitude higher than in both other regimens. It is historically known that enterococci tend to increase in numbers under similar use of topical antibiotics [35], as these Gram-positive bacteria are naturally resistant to the SDD-antibiotics. Also, Gram-negative intestinal bacteria induce an antimicrobial peptide, Reg3g, at the luminal surface of intestinal cells by stimulating TLR4 [36], with growth limiting effects on enterococci. Elimination of Gram negative bacteria resulted in a decrease of Reg3g with subsequent increase and translocation of enterococci in a mouse model [36].
The enterococci were considered to be harmless when SDD was first introduced in the ICU and none of the included patients had bloodstream infections with enterococci. However, we are aware that enterococci cause serious nosocomial infections, spread easily, and acquire increased antibiotic resistance [37, 38]. Enterococci are the third leading cause of endocarditis, and nosocomial acquisition is associated with a poor prognosis [39, 40]. Furthermore, antibiotic resistance gene-transfer has been demonstrated in vivo between enterococci and other bacterial species [41].
Limitations to the study are the limited numbers of patients and samples. Also, by design of the study, no baseline samples could be obtained nor could the timing of the sampling be standardized, as all samples were produced spontaneously. Based upon the data provided in table 2 we believe this has not caused a major source of bias.
Conclusion
We show that the total numbers of bacteria of the faecal microbiota in patients in the ICU are not significantly influenced by SDD. Enterobacteriaceae dropped significantly in numbers in the SDD regimen compared to SC and SOD regimens, as expected.
SDD does have a significant impact on the composition of the anaerobic intestinal microbiota; the number of _F. prausnitzii_-group of bacteria is significantly reduced during the SDD regimen compared to both SC and SOD regimens. The bacteria from the Enterococcus groups are present in significantly higher numbers in the SDD compared to both SC and SOD regimens. The other groups of bacteria show some variations but these are not significant between both other regimens.
The hypothesis that SDD is unequivocally beneficial by only reducing Enterobacteriaceae while leaving the colonic microbiota unaffected has to be rejected.
References
- van der Waaij D, Berghuis-de Vries JM, Lekkerkerk-van der Wees JE (1971) Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond) 69:405–411
Google Scholar - van der Waaij D (1992) History of recognition and measurement of colonization resistance of the digestive tract as an introduction to selective gastrointestinal decontamination. Epidemiol Infect 109:315–326
Article PubMed Google Scholar - Stoutenbeek CP, van Saene HK, Miranda DR, Zandstra DF (1984) The effect of selective decontamination of the digestive tract on colonisation and infection rate in multiple trauma patients. Intensive Care Med 10:185–192
Article PubMed CAS Google Scholar - D’Amico R, Pifferi S, Leonetti C, Torri V, Tinazzi A, Liberati A (1998) Effectiveness of antibiotic prophylaxis in critically ill adult patients: systematic review of randomised controlled trials. BMJ 316:1275–1285
PubMed Google Scholar - Liberati A, D’Amico R, Pifferi, Torri V, Brazzi L (2004) Antibiotic prophylaxis to reduce respiratory tract infections and mortality in adults receiving intensive care. Cochrane Database Syst Rev (CD000022)
- Vandenbroucke-Grauls CM, Vandenbroucke JP (1991) Effect of selective decontamination of the digestive tract on respiratory tract infections and mortality in the intensive care unit. Lancet 338:859–862
Article PubMed CAS Google Scholar - Krueger WA, Lenhart FP, Neeser G, Ruckdeschel G, Schreckhase H, Eissner HJ, Forst H, Eckart J, Peter K, Unertl KE (2002) Influence of combined intravenous and topical antibiotic prophylaxis on the incidence of infections, organ dysfunctions, and mortality in critically ill surgical patients: a prospective, stratified, randomized, double-blind, placebo-controlled clinical trial. Am J Respir Crit Care Med 166:1029–1037
Article PubMed Google Scholar - de Jonge E, Schultz MJ, Spanjaard L, Bossuyt PM, Vroom MB, Dankert J, Kesecioglu J (2003) Effects of selective decontamination of digestive tract on mortality and acquisition of resistant bacteria in intensive care: a randomised controlled trial. Lancet 362:1011–1016
Article PubMed CAS Google Scholar - Bergmans DC, Bonten MJ, Gaillard CA, Paling JC, van der Geest S, van Tiel FH, Beysens AJ, de Leeuw PW, Stobberingh EE (2001) Prevention of ventilator-associated pneumonia by oral decontamination: a prospective, randomized, double-blind, placebo-controlled study. Am J Respir Crit Care Med 164:382–388
PubMed CAS Google Scholar - Liberati A, D’Amico R, Pifferi S, Torri V, Brazzi L, Gensini GF, Gusinu R (2006) Antibiotic prophylaxis to prevent nosocomial infections in patients in intensive care units: evidence that struggle to convince practising clinicians. Intern Emerg Med 1:160–162
Article PubMed Google Scholar - Koeman M, van der Ven AJ, Hak E, Joore HC, Kaasjager K, de Smet AG, Ramsay G, Dormans TP, Aarts LP, de Bel EE, Hustinx WN, van der Tweel I, Hoepelman AM, Bonten MJ (2006) Oral decontamination with chlorhexidine reduces the incidence of ventilator-associated pneumonia. Am J Respir Crit Care Med 173:1348–1355
Article PubMed CAS Google Scholar - de Smet AM, Kluytmans JA, Cooper BS, Mascini EM, Benus RF, van der Werf TS, van der Hoeven JG, Pickkers P, Bogaers-Hofman D, van der Meer NJ, Bernards AT, Kuijper EJ, Joore JC, Leverstein-van Hall MA, Bindels AJ, Jansz AR, Wesselink RM, de Jongh BM, Dennesen PJ, van Asselt GJ, te Velde LF, Frenay IH, Kaasjager K, Bosch FH, van Iterson M, Thijsen SF, Kluge GH, Pauw W, de Vries JW, Kaan JA, Arends JP, Aarts LP, Sturm PD, Harinck HI, Voss A, Uijtendaal EV, Blok HE, Thieme Groen ES, Pouw ME, Kalkman CJ, Bonten MJ (2009) Decontamination of the digestive tract and oropharynx in ICU patients. N Engl J Med 360:20–31
Article PubMed Google Scholar - Vanhoutte T, Huys G, de Brandt E, Swings J (2004) Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol Ecol 48:437–446
Article PubMed CAS Google Scholar - Gibson GR, Roberfroid MB (1995) Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 125:1401–1412
PubMed CAS Google Scholar - van der Waaij D, Manson WL, Arends JP, de Vries-Hospers HG (1990) Clinical use of selective decontamination: the concept. Intensive Care Med 16(Suppl 3):S212–S216
Article PubMed Google Scholar - Harmsen HJ, Gibson GR, Elfferich P, Raangs GC, Wildeboer-Veloo AC, Argaiz A, Roberfroid MB, Welling GW (2000) Comparison of viable cell counts and fluorescence in situ hybridization using specific rRNA-based probes for the quantification of human fecal bacteria. FEMS Microbiol Lett 183:125–129
Article PubMed CAS Google Scholar - Harmsen HJ, Raangs GC, He T, Degener JE, Welling GW (2002) Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol 68:2982–2990
Article PubMed CAS Google Scholar - Knaus WA, Draper EA, Wagner DP, Zimmerman JE (1985) APACHE II: a severity of disease classification system. Crit Care Med 13:818–829
Article PubMed CAS Google Scholar - Franks AH, Harmsen HJ, Raangs GC, Jansen GJ, Schut F, Welling GW (1998) Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol 64:3336–3345
PubMed CAS Google Scholar - Poulsen LK, Lan F, Kristensen CS, Hobolth P, Molin S, Krogfelt KA (1994) Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun 62:5191–5194
PubMed CAS Google Scholar - Waar K, Degener JE, van Luyn MJ, Harmsen HJ (2005) Fluorescent in situ hybridization with specific DNA probes offers adequate detection of Enterococcus faecalis and Enterococcus faecium in clinical samples. J Med Microbiol 54:937–944
Article PubMed CAS Google Scholar - Jansen GJ, Wildeboer-Veloo AC, Tonk RH, Franks AH, Welling GW (1999) Development and validation of an automated, microscopy-based method for enumeration of groups of intestinal bacteria. J Microbiol Methods 37:215–221
Article PubMed CAS Google Scholar - Suau A, Rochet V, Sghir A, Gramet G, Brewaeys S, Sutren M, Rigottier-Gois L, Doré J (2001) Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst Appl Microbiol 24:139–145
Article PubMed CAS Google Scholar - Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105:16731–16736
Article PubMed CAS Google Scholar - Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ (2002) Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol 52:2141–2146
Article PubMed CAS Google Scholar - Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ (2002) The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217:133–139
Article PubMed CAS Google Scholar - Koruda MJ, Rolandelli RH, Bliss DZ, Hastings J, Rombeau JL, Settle RG (1990) Parenteral nutrition supplemented with short-chain fatty acids: effect on the small-bowel mucosa in normal rats. Am J Clin Nutr 51:685–689
PubMed CAS Google Scholar - Whelan K, Judd PA, Preedy VR, Simmering R, Jann A, Taylor MA (2005) Fructooligosaccharides and fiber partially prevent the alterations in fecal microbiota and short-chain fatty acid concentrations caused by standard enteral formula in healthy humans. J Nutr 135:1896–1902
PubMed CAS Google Scholar - Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE (2007) Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 73:1073–1078
Article PubMed CAS Google Scholar - van Saene HK, Stoutenbeek CP, Zandstra DF (1988) Cefotaxime combined with selective decontamination in long term intensive care unit patients. Virtual absence of emergence of resistance. Drugs 35 Suppl 2:29–34
Article PubMed Google Scholar - Vollaard EJ, Clasener HA, Janssen AJ, Wynne HJ (1990) Influence of cefotaxime on microbial colonization resistance in healthy volunteers. J Antimicrob Chemother 26:117–123
Article PubMed CAS Google Scholar - Guggenbichler JP, Kofler J, Allerberger F (1985) The influence of third-generation cephalosporins on the aerobic intestinal flora. Infection 13(Suppl 1):S137–S139
Article PubMed Google Scholar - Welling GW, Groen G (1993) Specific inactivation of antimicrobial agents and its interindividual differences. Old Herborn University Seminar Monographs 3. The Institute for Microbiology and Biochemistry, Herborn-Dill, pp 47–54
Google Scholar - Pultz NJ, Stiefel U, Subramanyan S, Helfand MS, Donskey CJ (2005) Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. J Infect Dis 191:949–956
Article PubMed Google Scholar - Bonten MJ, Gaillard CA, van Tiel FH, van der GS, Stobberingh EE (1995) Colonization and infection with Enterococcus faecalis in intensive care units: the role of antimicrobial agents. Antimicrob Agents Chemother 39:2783–2786
- Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG (2008) Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455:804–807
Article PubMed CAS Google Scholar - Montecalvo MA, Horowitz H, Gedris C, Carbonaro C, Tenover FC, Issah A, Cook P, Wormser GP (1994) Outbreak of vancomycin-, ampicillin-, and aminoglycoside-resistant Enterococcus faecium bacteremia in an adult oncology unit. Antimicrob Agents Chemother 38:1363–1367
PubMed CAS Google Scholar - Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJ (2005) Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis 11:821–828
PubMed CAS Google Scholar - Fernandez Guerrero ML, Goyenechea A, Verdejo C, Roblas RF, de GM (2007) Enterococcal endocarditis on native and prosthetic valves: a review of clinical and prognostic factors with emphasis on hospital-acquired infections as a major determinant of outcome. Medicine (Baltimore) 86:363–377
- Stevens MP, Edmond MB (2005) Endocarditis due to vancomycin-resistant enterococci: case report and review of the literature. Clin Infect Dis 41:1134–1142
Article PubMed Google Scholar - Netherwood T, Bowden R, Harrison P, O’Donnell AG, Parker DS, Gilbert HJ (1999) Gene transfer in the gastrointestinal tract. Appl Environ Microbiol 65:5139–5141
PubMed CAS Google Scholar
Acknowledgments
R.F.J. Benus has received a grant from ‘UMCG Stimuleringsgelden’. We thank M. Tanweer Khan for the culturing of F. prausnitzii and performing the MIC-tests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
- Department of Medical Microbiology, University Medical Centre Groningen, University of Groningen, Groningen, The Netherlands
Robin F. Benus, Hermie J. Harmsen, Gjalt W. Welling & John E. Degener - Department of Critical Care, University Medical Centre Groningen, University of Groningen, Groningen, The Netherlands
Robin F. Benus, Rob Spanjersberg & Jan G. Zijlstra - Infectious Diseases and Tuberculosis Service, Department of Internal Medicine, Pulmonary Diseases and Tuberculosis, University Medical Centre Groningen, University of Groningen, Groningen, The Netherlands
Robin F. Benus & Tjip S. van der Werf - P.O. Box 30001, Hanzeplein 1, AA11, room U3.105, 9700 RB, Groningen, The Netherlands
Tjip S. van der Werf
Authors
- Robin F. Benus
- Hermie J. Harmsen
- Gjalt W. Welling
- Rob Spanjersberg
- Jan G. Zijlstra
- John E. Degener
- Tjip S. van der Werf
Corresponding author
Correspondence toTjip S. van der Werf.
Electronic supplementary material
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Benus, R.F., Harmsen, H.J., Welling, G.W. et al. Impact of digestive and oropharyngeal decontamination on the intestinal microbiota in ICU patients.Intensive Care Med 36, 1394–1402 (2010). https://doi.org/10.1007/s00134-010-1826-4
- Received: 13 August 2009
- Accepted: 14 January 2010
- Published: 16 March 2010
- Issue date: August 2010
- DOI: https://doi.org/10.1007/s00134-010-1826-4