FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions (original) (raw)
Introduction
Frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) share many pathological, genetic, and molecular features [23, 25, 32]. Indeed, in both disorders, GGGGCC (G4C2) repeat expansion in the noncoding region of C9ORF72 is the most common genetic mutation [10, 16, 31]. G4C2 sense and antisense transcripts form RNA foci in patient brains, fibroblasts, induced pluripotent stem cells (iPSCs), and iPSC-derived neurons [1, 10, 13, 15, 22, 27, 34, 42], which may contribute to disease pathogenesis through an RNA-mediated mechanism. For instance, some RNA-binding proteins are sequestered in these foci [29]. However, it is not known which of these proteins shows compromised function that may play a key pathogenic role. On the other hand, abnormal dipeptide repeat proteins (DPRs) arising from repeat-associated non-AUG (RAN) translation—a process discovered in spinocerebellar ataxia type 8 and myotonic dystrophy type 1 [41]—form mostly cytoplasmic inclusions in the brain and spinal cord of patients with C9ORF72 repeat expansion [3, 30, 42]. Six DPRs can be translated from both sense and antisense expansion transcripts and adjacent intronic sequences of C9ORF72 [9]. However, the specific roles and mechanisms of each DPR in disease pathogenesis in vivo are largely unknown.
To address these important questions, we generated DNA constructs containing 80 copies of GGXGCX, GGXCGX, or CCXCGX (with X being randomly any one of the four nucleotides) and corresponding transgenic flies that express individual DPRs, such as (GA)80, (GR)80, and (PR)80. When expressed in a cell-type-specific manner, (GR)80 and (PR)80, but not (GA)80, were toxic in neuronal and non-neuronal cells in Drosophila. In contrast to a previous report [21], (GR)80 was mostly localized throughout the cytosol and did not accumulate in the nucleolus in neurons and wing disc cells. (GR)80 suppressed Notch signaling, resulting in cell loss in the wing, a phenotype that was largely suppressed by ectopic expression of Notch. Consistent with these findings, we found lower expression levels of some Notch target genes in iPSC-derived cortical neurons and brain tissues of C9ORF72 patients. (GR)80 toxicity was partially suppressed by co-expression of (GA)80, which recruited (GR)80 into cytoplasmic inclusions in Drosophila cells, HeLa cells, and iPSC-derived human neurons. Thus, the Notch pathway, one of many receptor signaling pathways, is a target of poly(GR) toxicity that in turn can be suppressed by poly(GA) likely through inclusion formation.
Materials and methods
Generation and maintenance of transgenic fly lines
Artificial DNA sequences were designed to express 80 repeats of GA, GR, and PR following a Flag tag at the N-terminus. As a control, a stop codon TAA was introduced instead of the ATG codon at the beginning of the open reading frame of GA. The DNA sequences were synthesized (Genewiz) and subcloned into pUASTattB vector between the Bgl_II and Xho_I sites. The UAS-(GR)80 control construct was generated by site-directed mutagenesis (QuikChange II Site-Directed Mutagenesis Kits, Agilent Technologies), in which the first two nucleotides of the start codon ATG were mutated to TA to form the stop codon TAG (the a5263t_t5264a mutations primer: TCGTTAACAGATCTCCAC CTAGGATTACAAGGACGACGAC). Transgenic flies were made with transgenes UAS_-(GA) 80 control, UAS_-(GA) 80 , UAS_-(GR)_ 80 control, UAS_-(GR)_ 80 , and _UAS_-(PR) 80 present on the second or third chromosome. The presence of transgenes in the fly was confirmed by sequencing the genomic region amplified by RT-PCR. Primers used were CTGCAACTACTGAAATCTGCCA (forward) and TGTCACACCACAGAAGTAAGGT (reverse). CTGCAACTACTGAAATCTGCCA (forward) was used for sequencing. All flies were raised at 25 °C on a standard diet. _GMR_-Gal4, _OK371_-Gal4, _UAS_-GFP/CyO, _Vg_-Gal4, and w 1118; _P{NRE_-EGFP.S}5A flies were from the Bloomington Drosophila Stock Center. Notch 5419 mutant and _UAS_-Notch FL7 flies were kindly provided by Dr. S. Artavanis-Tsakonas.
Quantifying the adult wing notching phenotype
Wings were arbitrarily classified into four groups according to the strength of the phenotype (absent, weak, medium, and strong), as judged from the number of notches and the size of wing area lost.
Climbing assay and quantification of dendritic branching
UAS_-(GA)_ 80 control, UAS_-(GA)_ 80 , UAS_-(GR)_ 80 , and UAS-(PR) 80 flies were crossed with _OK371_-Gal4, _UAS_-GFP/CyO flies at 18 °C to obtain flies expressing dipeptides in motor neurons. For the climbing assay, individual 3-day-old adult flies were placed into a 15-ml polypropylene centrifuge tube (CellTreat Scientific Products). After 1 min, each fly was lightly taped to the bottom of the tube and allowed to climb for 10 s. The climbing distance was scored as the average of five tests for each fly. For quantification of dendritic branching, ddaE sensory neurons were labeled with mCD8-GFP driven by _221_-Gal4. The number of dendritic ends was counted at the third-instar larval stage even though the mCD8-GFP signal was reduced by (GR)80 and (PR)80.
Immunohistochemistry
Third-instar larval wing imaginal discs, brains or salivary glands were dissected in PBS and fixed in 4 % paraformaldehyde for 20 min at room temperature. After three washes in PBS, samples were permeabilized in PBS containing 0.5 % Triton X-100 (PBT) for 30 min at room temperature and then blocked in 0.5 % goat serum in PBT for 1 h at room temperature. Samples were then incubated with the primary antibody overnight at 4 °C, washed three times with PBT, and incubated with secondary antibody for 2 h at room temperature. The primary antibodies were mouse anti-Flag (Sigma; 1:500) and mouse anti-Wingless (Wg) (Developmental Studies Hybridoma Bank; 1:50). The secondary antibodies were goat anti-mouse Alexa 488 and Alexa 594 (Invitrogen; 1:200). DNA staining was carried out by incubating salivary glands with Quant-iT OliGreen ssDNA reagent (Life Technologies; 1:1000) at room temperature for 5 min. Alternatively, samples were loaded on slides with Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI, Vector laboratories).
Neuronal cultures and human brain samples
Published iPSC lines from two control subjects and three C9ORF72 carriers [1, 2, 34] were differentiated into cortical neurons as described [2]. Neuronal cultures were aged for 8 weeks before RNA extraction. For (GA)80 and (GR)80 expression experiments, neuronal cultures from one control line were aged for 4 weeks before transfection. Middle frontal gyrus brain samples from three healthy control subjects and eight C9ORF72 repeat expansion carriers were obtained from the UCSF Memory and Aging Center. Another three control subjects were from the Mayo Clinic Jacksonville and were used in a recent study [14]. The mean age at death was 78 ± 8 years in six control subjects and 64 ± 7 years in eight C9ORF72 repeat expansion carriers.
Quantitative RT-PCR
Total RNA from patient neurons and brain tissues was extracted with the RNeasy kit (Qiagen) according to the manufacturer’s instructions. Total RNA (1000 ng) was reverse transcribed to cDNA with random hexamers and TaqMan reverse transcription reagents (Applied Biosystems). Real-time quantitative PCR was performed with SYBR Green Select Master Mix (Applied Biosystems) on a StepOnePlus system (Applied Biosystems); cyclophilin was used as internal control. Primers used were CGGACATTCTGGAAATGACA (HES1 forward), CATTGATCTGGGTCATGCAG (HES1 reverse), TATCGGAGTTTGGGATTTCG (HEY1 forward), GGGTGATGTCCGAAGACG (HEY1 reverse), TGCCATCGCCAAGGAGTAG (cyclophilin forward), TGCACAGACGGTCACTCAAA (Cyclophilin reverse), cctggatgactcttgggaaa (NFKB1 forward), and tcagccagctgtttcatgtc (NFKB1 reverse). For DPR mRNA analysis, total RNAs were extracted and purified from third-instar larvae tissue with the RNeasy Mini Kit. Ct values for each gene were normalized to Actin42A (primers are 5′-TCTTACTGAGCGCGGTTACAG-3′ and 5′-ATGTCGCGCACAATTTCAC-3′). Relative mRNA expression was calculated using the delta–delta Ct method. One pair of primers targeting UAST-attB 3′ UTR (5′-TTCCAACCTATGGAACTGATGA-3′ and 5′-GGTTTTCCTCATTA AAGGCATTC-3′) was selected to detect different dipeptide transcripts.
Transfection of neurons and HeLa cells and immunofluorescence
For transfection of 4-week-old iPSC-derived neurons and HeLa cells, we used 1.6 µg of plasmid DNA containing HA-(GA)80, FLAG-(GR)80, or empty vector (pcDNA 3.1, Life Technologies) and Lipofectamine 2000 (Life Technologies). Forty-eight hours after transfection, cells were fixed with 4 % paraformaldehyde and permeabilized with 0.2 % Triton X-100, blocked with 3 % bovine serum albumin, and incubated overnight at 4 °C with primary antibodies against HA (Roche; 1:600) or FLAG (Sigma; 1:1000). After three washes with PBS, the cells were incubated with Alexa Fluor-conjugated secondary antibodies (Invitrogen; 1:300) for 1 h at room temperature and mounted on glass slides with Vectashield HardSet Mounting Medium with DAPI. Immunostained cells were examined by fluorescence microscopy.
Imaging
The images of adult eyes and wings were acquired with a Nikon DS-Fi1 camera and a Nikon SMZ1500 stereomicroscope. The image of the dorsoventral boundary was acquired with a Nikon D-Eclipse C1. All other images were acquired with a Leica TCS SP5 II laser scanning confocal microscope and Leica LAS AF software. To quantify Wg fluorescence intensity in wing imaginal discs after staining, 30 Z-stack images (step size 0.5 μM) were acquired for each sample. Image-J was used for Z projection, and the integrated intensity of the same region at the dorsoventral boundary of six discs was measured per genotype.
Results
Generation of transgenic Drosophila models of DPR toxicity
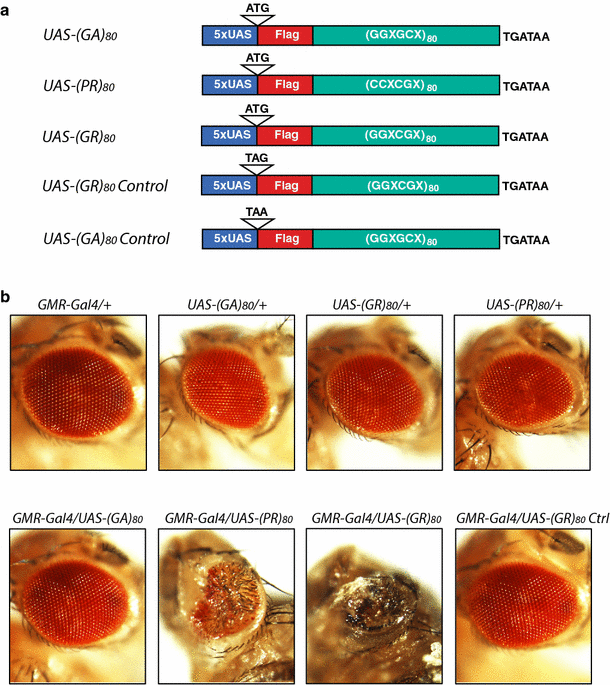
To determine which DPR is toxic in vivo and identify the mechanisms, we generated DNA constructs containing 80 copies of GGXGCX, GGXCGX, or CCXCGX (with X being randomly any one of the four nucleotides) that can be transcribed under the control of UAS elements and translated into (GA)80, (GR)80, or (PR)80, respectively. All constructs contained the CCACC consensus Kozak sequence adjacent to the ATG start codon and a DNA sequence encoding the Flag tag at the N-terminus of DPRs (Fig. 1a; Tab. S1). To generate transgenic fly lines, we used the PhiC31 integrase-mediated site-specific integration system [5] to ensure equal transcription of different DPR constructs in a cell-type specific manner when a unique Gal4 driver is used [8].
Fig. 1
Toxicity of different FTD/ALS-associated dipeptides in the Drosophila eye. a Schematic representation of DNA constructs that express Flag-tagged (GA)80, (PR)80, and (GR)80 under the control of the UAS elements. In control lines, the AUG start codon was replaced by a UAG or UAA stop codon to block translation. X at the third codon can be randomly any one of the four nucleotides. b Representative images of Drosophila eyes with different genotypes. Neither _GMR_-Gal4 nor any of the UAS transgenic lines showed an eye phenotype. Expression of (PR)80 or (GR)80 in the eye by the _GMR_-Gal4 resulted in grossly deformed eyes
To dissect the toxicity of DPRs, we first used _GMR_-Gal4, which drives ectopic gene expression mostly in the eye, a widely used model for studying neurodegeneration [6, 24]. Expression of (PR)80 and (GR)80, but not (GA)80, driven by _GMR_-Gal4 resulted in a mostly adult lethal phenotype and caused a drastic deformation of eyes in all flies at the pupal stage; neither the _GMR_-Gal4 nor any of the UAS transgenic lines alone had any eye phenotype (Fig. 1b). More than 200 flies were examined for each genotype. To confirm that different DPRs were expressed in the eye tissue, we performed RT-PCR analysis using primers specific to the 3′UTR sequence common to all DPR constructs. Indeed, all DPR mRNAs were expressed, although at different levels (Fig. S1), which is probably due to differences in mRNA stability. The stability, degradation, and biophysical properties of (GA)80 proteins may also differ from those of (PR)80 and (GR)80. Indeed, immunostaining analysis with Flag tag-specific antibody showed all three DPRs were expressed when _OK371_-Gal4 was used (Fig. S2a–c), which drives target gene expression in cholinergic neurons including motor neurons. However, unlike (GR)80 (Fig. S2b) and (PR)80 (Fig. S2c), (GA)80 forms distinct inclusions, mostly one in each neuron (Fig. S2a). Thus, although the extent of toxicity between different DPRs cannot be compared directly, lack of (GA)80 toxicity is correlated with its inclusion formation, while more diffuse (GR)80 and (PR)80 cause drastic phenotypes under the same experimental condition.
Fig. 2
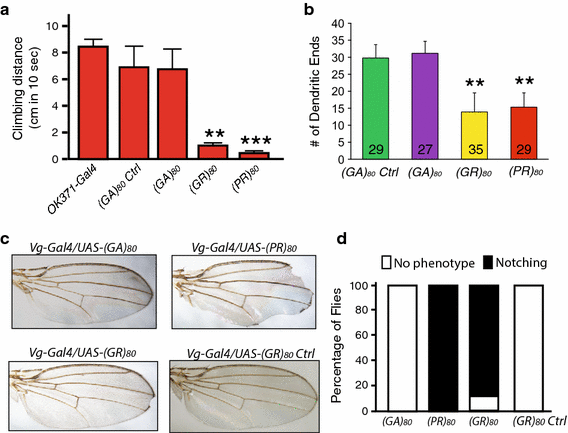
(GR)80 is toxic in neuronal and non-neuronal cells in Drosophila. a Effects of (GA)80, (GR)80, and (PR)80 on locomotor activity of adult flies. (GA)80, (GR)80, and (PR)80 were expressed in motor neurons by _OK371_-Gal4 at 18 °C, and surviving 3-day-old adult flies (10 flies of each genotype) were tested for climbing activity. Forty _OK371_-Gal4 flies and 10 flies expressing (GA) 80 mRNA but with a stop codon instead of the ATG start codon (see Table S1) were also examined. Values are mean ± SEM. **p value <0.01, ***p value <0.001, by Student’s t test. b Effects of (GA)80, (GR)80, and (PR)80 expression on dendritic branching of ddaE sensory neurons. ddaE neurons were labeled with mCD8-GFP driven by 221-Gal4. The number of neuron analyzed for each genotype is listed on each column. Values are mean ± SD. **p value <0.01, by single-factor ANOVA. c Effects of (GA)80, (GR)80, and (PR)80 on the survival of wing margin cells. The expression of (GR)80, and (PR)80 resulted in the wing notching phenotype. d The percentage of flies with or without wing notching phenotype is shown for each genotype. More than 300 flies of both sexes were scored for each genotype
To further exclude the possibility that (GR)80 toxicity might be caused by transcription of GC-rich RNAs in vivo, we also generated transgenic fly lines that expressed the same (GR) 80 mRNA except that the AUG start codon was replaced with the UAG stop codon (Fig. 1a). Expression of this control construct from the same genomic locus under the same Gal4 driver was not toxic (Fig. 1b). Thus, (GR)80 and (PR)80 proteins are highly toxic when overexpressed in the Drosophila eye by the _GMR_-Gal4.
(GR)80 is toxic in neuronal and non-neuronal cells in Drosophila
Since _GMR_-Gal4 drives target gene expression at a high level, the drastic toxicity of (GR)80 and (PR)80 in the eye should be considered in the context of their overexpression. Therefore, we searched for Gal4 drivers that would give rise to a more moderate phenotype, enabling us to study the mechanism. Expression of the (GA)80 construct by various Gal4s did not result in any obvious phenotypes (Table S2). However, expression of (GR)80 and (PR)80 in all cells by _tubulin_-Gal4, in all neurons by _elav_-Gal4, or in motor neurons by D42 or _OK371_-Gal4 at 25 °C resulted in a mostly lethal phenotype (Table S2). The strength of Gal4 drivers is dependent on temperature. Lower expression of (GR)80 and (PR)80 by _OK371_-Gal4 at 18 °C caused a semi-lethal phenotype (Table S2), and surviving adult flies had greatly reduced locomotor activity (Fig. 2a). Moreover, expression of (GR)80 and (PR)80 in ddaE sensory neurons decreased dendritic branching (Fig. 2b). (GR)80-control was not used in these experiments because it did not produce any eye phenotype (Fig. 1b). Expression of (GR)80, but not the (GR)80 control construct, by _Vg_-Gal4, which drives gene expression in the dorsoventral boundary and some other cells of the wing imaginal discs [11], gave rise to wing margin defects in 90 % of flies of both sexes at 25 °C (Fig. 2c, d). Thus, (GR)80 is toxic in multiple neuronal and non-neuronal cell types in vivo.
(GR)80 genetically interacts with Notch
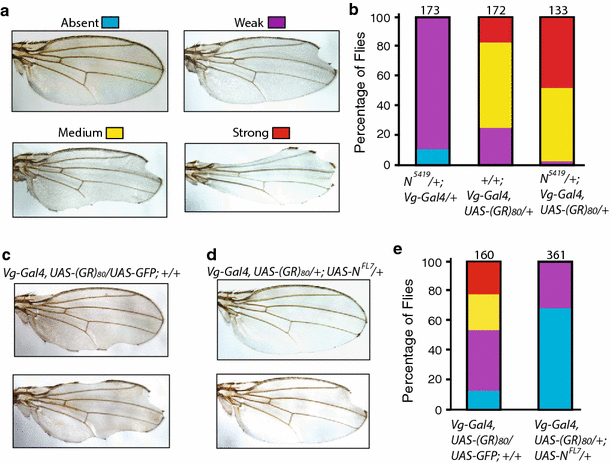
Misregulation of several signaling pathways results in distinct wing defects [4]. However, the mild notching defects at or near the tip of the wing caused by (GR)80 are remarkably similar to the defect due to partial loss of Notch activity, as in flies heterozygous for the N 5419 allele [20]. Therefore, we investigated the genetic interaction between Notch and (GR)80. We grouped wing notching phenotypes by their severity: absent, weak, medium, and strong (Fig. 3a). Since the Notch gene is located on the X chromosome, we examined only female flies in this genetic interaction experiment. About 90 % of female flies heterozygous for the N 5419 allele had a weak wing margin defect (Fig. 3b). To facilitate the genetic interaction study, Vg_-Gal4 and UAS_-(GR) 80 were recombined onto the same second chromosome. The wing notching phenotype in the resulting fly line (Fig. 3b) was stronger than that in flies transheterozygous for Vg_-Gal4 and UAS_-(GR) 80 (Fig. 2c). When this line was examined in the N 5419 _/_+ background, the wing notching phenotype was substantially stronger than would be expected from an additive effect of (GR)80 and N 5419 /+: 18 % of (GR)80 flies versus 47 % of (GR)80 flies in the N 5419 _/_+ background showed a strong wing margin phenotype (p < 0.05, n = 3), raising the possibility that the two genes genetically interact (Fig. 3b). These results suggest that (GR)80 compromises the Notch signaling pathway. Indeed, (GR)80 toxicity was largely suppressed by ectopic expression of full-length Notch; a wing notching phenotype was absent in 67 % of (GR) 80 flies when Notch was co-expressed but in only 12 % of (GR) 80 flies when GFP was co-expressed (p < 0.001, n = 3) (Fig. 3c–e). (PR)80 toxicity was not suppressed by Notch expression, and PR80 expression in N 5419 _/_+ background did not significantly enhance the PR80 wing defect (not shown), suggesting divergent pathogenic mechanisms of these two DPRs. To ensure that the suppression by Notch was not due to the dilution of Gal4 caused by a second copy of the UAS transgene, flies expressing both (GR)80 and GFP by _Vg_-Gal4 were used as controls in this experiment (Fig. 3c). These genetic analyses suggest that the Notch pathway is a major target of (GR)80 toxicity in vivo.
Fig. 3
Notch expression suppresses (GR)80 toxicity. a Representative images of Drosophila wings with margin defects of different severities. The image of a normal wing was from a UAS_-(GR)_ 80 /+ fly; other wing images were from N 5419/+; Vg_-Gal4, UAS_-(GR) 80 /+ flies. b Quantification of wing margin defects in female flies heterozygous for the N 5419 allele, expressing (GR) 80 driven by _Vg_-Gal4 recombined to the same second chromosome and expressing (GR) 80 on the N 5419/+ background by _Vg_-Gal4. Only female flies were examined, and the total number of flies for each genotype from three experiments is listed above each column. c Representative images of wing margin defects in Vg_-Gal4, UAS_-(GR) 80 _/_+ flies. d Representative wing images of Vg_-Gal4, UAS_-(GR) 80 /+; _UAS_-N FL7 flies showing suppression of the wing margin defects by ectopic expression of full-length Notch. _UAS_-GFP was used as the control for _UAS_-N FL7. e Quantification of Notch suppression of wing margin defects caused by (GR)80. The total number of flies of both sexes examined for each genotype from 3 experiments is listed above each column
(GR)80 suppresses Notch signaling
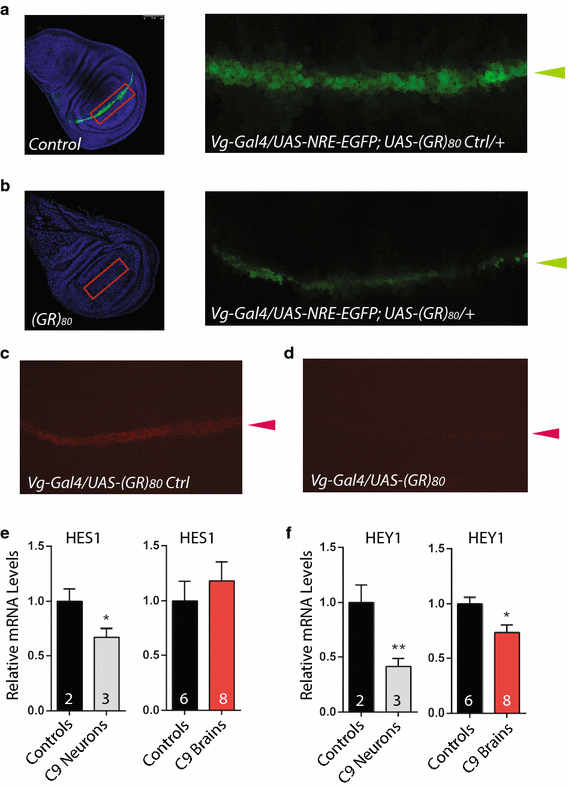
To confirm (GR)80 indeed suppresses Notch signaling, we expressed a Notch activity reporter, _UAS_-_Notch Response Element (NRE)-EGFP [33], in the wing disc cells by Vg_-Gal4. Notch signaling was generally lower in all cells at the dorsoventral boundary that express UAS_-(GR) 80 (Fig. 4b) than in control flies expressing UAS_-(GR) 80 Control (Fig. 4a). The dorsoventral boundary remained intact, and no obvious cell loss was observed, as judged from caspase-3 immunostaining. To further confirm this finding, we examined the expression level of endogenous Wingless (Wg) at the dorsoventral boundary of the wing disc, which is a direct target of Notch signaling [12]. Indeed, (GR)80 significantly suppressed endogenous Wg expression (Fig. 4c, d). Although we cannot rule out the possibility that (GR)80 affects the Wg level through other mechanisms, this result is consistent with the finding that (GR)80 decreases endogenous Notch signaling.
Fig. 4
(GR)80 downregulates notch signaling in Drosophila. GFP expression controlled by the Notch-responsive element (NRE) at the dorsoventral boundary (green arrowheads) of a control wing disc (a) and a wing disc expressing (GR)80 driven by _Vg_-Gal4 (b). The areas indicated by red rectangles are enlarged and presented as the two right panels. Wg immunostaining at the dorsoventral boundary (red arrowheads) of a control wing disc (c) and a wing disc expressing (GR)80 (d). Expression levels of the Notch targets HES1 (e) and HEY1 (f) in iPSC-derived neurons and brain tissues of subjects with C9ORF72 repeat expansion. The number of iPSC lines or brain samples analyzed is indicated in each column. Values are mean ± SEM. *p value <0.05, **p value <0.01 by Student’s t test
Poly(GR) proteins have been detected mostly in the cytoplasmic inclusions in brain tissues of patients with C9ORF72 repeat expansion [3, 30, 42], thus it is possible that Notch signaling in patient neurons may also be compromised. In recent years, patient-specific iPSCs have been used as a powerful model for many neurodegenerative diseases [18, 38, 39]. We differentiated iPSC lines from two control and three subjects with C9ORF72 repeat expansions [1, 2, 34] into 8-week-old postmitotic neurons of cortical lineage and found reduced expression of the Notch target genes HES1 (Fig. 4e) and HEY1 (Fig. 4f). HEY1, one of the target genes more sensitive to Notch level [35], showed reduced expression in brain tissues of C9ORF72 patients (Fig. 4f). The expression of Notch target NFKB1 was also lower in both iPSC-derived neurons and brain tissues of C9ORF72 patients (not shown). Although these results are correlative, Notch signaling seems to be compromised in patient cells as well.
(GA)80 partially suppresses (GR)80 toxicity through inclusion formation
To further examine the effects of (GR)80 on Notch signaling, we examined the subcellular localization of (GR)80 expressed in a subset of wing disc cells by _Vg_-Gal4 (Fig. S2d). Immunostaining with either anti-Flag antibody or anti-GR antibody revealed that this DPR was localized in the cytoplasm (Fig. S2e and g), suggesting that (GR)80 interferes with the Notch signaling pathway in the cytoplasm. In both wing disc cells (Fig. S2f) and salivary gland cells (Fig. S3d), (PR)80 also seemed to be mostly cytoplasmic.
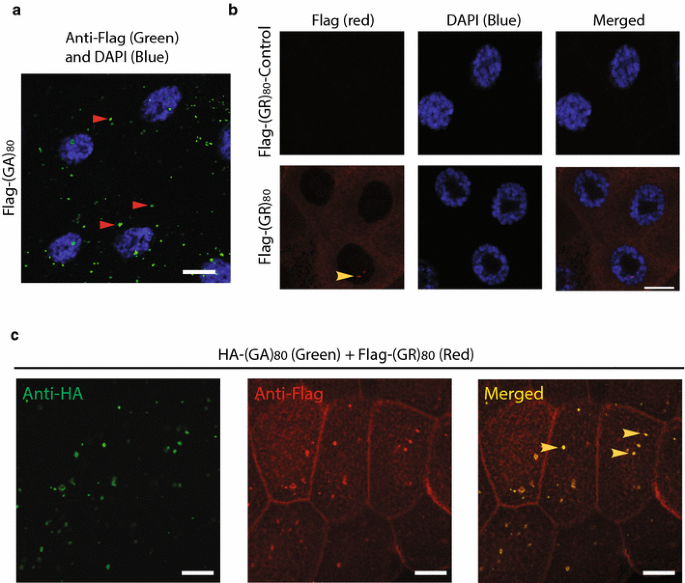
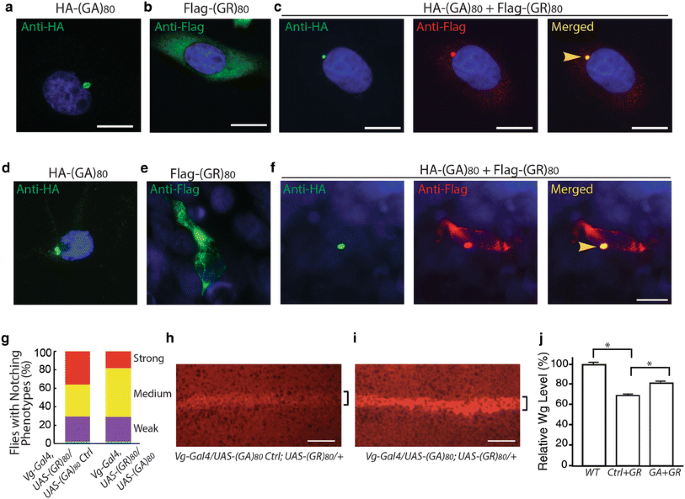
Since both poly(GA) and poly(GR) proteins are concomitantly expressed in patient cells [3, 30, 42], we first co-expressed (GA)80 and (GR)80 in Drosophila salivary gland cells, which are large and facilitate imaging analysis. (GA)80 by itself formed mostly cytoplasmic inclusions (Fig. 5a), while (GR)80 was localized throughout the cytoplasm in salivary gland cells (Fig. 5b), as in wing disc cells (Fig. S2e). This subcellular distribution of (GR)80 in salivary gland cells was confirmed with the anti-GR antibody (Fig. S3e). We also noticed one or two small (GR)80-positive dots on chromatin in each salivary gland cell (Fig. 5b, Fig. S3e), as well as in Drosophila motor neurons (Fig. S3f). Their nature and significance remain to be determined. Shorter GR forms have been reported to accumulate in nucleoli of cultured cells [21], but we found no significant presence of (GR)80 in nucleoli of salivary gland cells, even though their nucleoli were enlarged (Fig. 5b). But unexpectedly, in the presence of HA-(GA)80 (Fig. 5c, left panel), a portion of Flag-(GR)80 formed cytoplasmic inclusions as well (Fig. 5c, middle panel), in contrast to the diffuse cytoplasmic localization when only Flag-(GR)80 was expressed (Fig. 5b). Because (GA)80 alone forms cytoplasmic inclusions (Fig. 5a), co-localization of Flag-(GR)80 and HA-(GA)80 suggested that (GA)80 recruits (GR)80 into these inclusions (Fig. 5c, right panel).
Fig. 5
Subcellular localization of (GR)80 and (GA)80. a Flag-tagged (GA)80 forms inclusions mostly in the cytosol of Drosophila salivary gland cells. Some inclusions are highlighted by red arrowheads. b (GR)80 is largely present throughout the cytoplasm of salivary gland cells, and their nucleoli are larger than those of cells expressing the (GR)80-control construct. These are confocal images, and one or two (GR)80-positive dots (yellow arrowhead) were observed on chromatin in each cell at different confocal planes. Scale bar 20 μm. c HA-tagged (GA)80 recruits Flag-tagged (GR)80 into cytoplasmic inclusions when the two are co-expressed. All inclusions contain both (GA)80 and (GR)80 (some are indicated by yellow arrowheads)
Similarly, in human HeLa cells, (GA)80 alone formed inclusions (Fig. 6a), while (GR)80 alone was localized throughout the cytoplasm without any detectable accumulation in the nucleus (Fig. 6b). (GA)80 inclusions seemed to be aggresomes surrounded by vimentin and were often close to the nucleus (Fig. S4a) and positive for p62 (Fig. S4b). When (GA)80 and (GR)80 were co-expressed, some (GR)80 co-localized with (GA)80 inclusions in all HeLa cells we examined (Fig. 6c). Moreover, in iPSC-derived human neurons, (GA)80 recruited (GR)80 into cytoplasmic inclusions as well (Fig. 6d–f). These findings raise the possibility that (GA)80 protein is protective and can sequester highly toxic poly(GR) protein into inclusions. Indeed, (GA)80 expression partially suppressed the (GR)80-induced cell-loss phenotype at the wing margin (Fig. 6g) and correspondingly increased the Notch signaling, as indicated by the elevated expression of Wg at the dorsoventral boundary of the wing disc (Fig. 6h–j). (GA)80 expression did not suppress the (GR)80-induced eye phenotype (not shown), presumably because _GMR_-Gal4 is a very strong driver so that the level of non-aggregated (GR)80 remained high. Thus, (GA)80 partially suppresses (GR)80 toxicity in vivo by sequestering (GR)80 into inclusions.
Fig. 6
(GA)80 suppresses (GR)80 toxicity through inclusion formation. a HA-tagged (GA)80 forms inclusion in HeLa cells. b (GR)80 expression alone in HeLa cells shows diffuse cytoplasmic localization. c When (GA)80 and (GR)80 are co-expressed in HeLa cells, (GR)80 is recruited into (GA)80 inclusions (yellow arrowhead). d HA-tagged (GA)80 forms inclusion in iPSC-derived human neurons. e (GR)80 expression alone in iPSC-derived human neurons shows cytoplasmic localization. f When (GA)80 and (GR)80 are co-expressed in iPSC-derived human neurons, (GR)80 is recruited into (GA)80 inclusions (yellow arrowhead). Scale bar 10 μm. g (GA)80 partially suppresses (GR)80 toxicity in wing disc cells, resulting in a less severe wing notching phenotype. In this experiment, Vg_-Gal4 and UAS_-(GR) 80 were recombined onto the same chromosome. Wg expression at the dorsoventral boundary of wing discs expressing UAS_-(GA)_ 80 control (Table S1) and UAS_-(GR)_ 80 (h) or both UAS_-(GA)_ 80 and UAS_-(GR)_ 80 (i). UAS_-(GR)_ 80 on the third chromosome. The brackets indicate the dorsoventral boundary. j Wg expression levels in wing discs of flies with genotypes described in panels h and i. Scale bar in panels a–f, h, i: 20 μm. Values are mean ± SEM. *p value <0.05 by single-factor ANOVA
Discussion
This study shows that poly(GR) proteins are toxic in various neuronal and non-neuronal cell types in vivo under defined experimental conditions, consistent with several reports published during the preparation of our manuscript [21, 28, 36, 37]. However, in contrast to some of these studies, in which smaller poly(GR) proteins provided to cultured cells migrate to and significantly accumulate in the nucleolus, we found that (GR)80 is mostly localized throughout the cytoplasm in vivo and does not accumulate in the nucleolus. Thus, it is possible that DPRs of different lengths have diverse biological properties and pathogenic mechanisms.
We found that (GR)80 compromises Notch signaling and that ectopic expression of Notch partially suppresses (GR)80 toxicity in Drosophila. The expression levels of DPRs in many experimental systems, including ours in this study, are likely much higher than in human neurons, especially before disease onset. Thus, detrimental effects of low levels of DPRs in postmitotic neurons over a long period remain to be examined, and some of the mechanisms may differ from those found in overexpression studies. Nonetheless, Notch signaling also seems to be downregulated in iPSC-derived neurons and postmortem brain tissues of subjects with C9ORF72 repeat expansion; however, this result is correlative and should be interpreted cautiously. Together, our results raise the possibility that some key receptor signaling pathways are compromised in C9ORF72 FTD/ALS patients. The Notch signaling pathway is subject to complex regulation at multiple steps [17]. It would be interesting to determine exactly how (GR)80 and possibly other poly(GR) proteins of different lengths affect the Notch pathway or other receptor signaling pathways. The relative contributions of the Notch pathway and other affected signaling pathways to dysfunction of patient neurons also remain to be examined.
Another pathological feature of (GR)80 was the presence of small (GR)80-positive dots on chromatin in postmitotic cells in Drosophila (Fig. 5b, Fig. S3e, f) but not in dividing HeLa cells (Fig. 6b). The nature and significance of these (GR)80-positive structures on chromatin are unknown. If similar structures can be found in diseased neurons of C9ORF72 FTD/ALS patients, our fly model would be a useful tool for further investigation. It is also worth noting that Drosophila cells expressing (GR)80 have enlarged nucleoli even in the absence of detectable accumulation of (GR)80 in this subcellular compartment. Various cellular stresses induce specific changes in nucleolar morphology and composition in many cell types, including postmitotic neurons [7, 19]. Thus, nucleolar defects in C9ORF72 FTD/ALS can be caused by indirect mechanisms other than direct actions of G4C2 repeat RNAs or DPRs in the nucleolus.
Subjects with C9ORF72 repeat expansion often live for decades without symptoms, and DPR inclusions are mostly cytoplasmic and more abundant in brain regions without neurodegeneration [3, 30, 42]. Our finding that (GA)80 suppresses (GR)80 toxicity by sequestering (GR)80 into cytoplasmic inclusions suggests that different DPRs have diverse roles in neuronal cell death. Although transient overexpression of poly(GA) alone in cultured cells can be detrimental [26, 40], our findings suggest that aggregation-prone poly(GA) proteins may also have an in vivo neuroprotective function in early stages of disease by sequestering highly toxic poly(GR) into inclusions. We speculate that in the early stage of disease, poly(GR) is efficiently sequestered by poly(GA) inclusions and degraded, likely through the autophagy pathway. But with disease progression, overall DPR toxicity in some neurons may increase due to elevated levels of non-aggregated poly(GR). Thus, sequestration of poly(GR) proteins and other approaches to alleviate their detrimental effects on receptor signaling pathways are potential therapeutic avenues.
References
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S et al (2013) Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol 126:385–399
Article PubMed Central CAS PubMed Google Scholar - Almeida S, Zhang Z, Coppola G, Mao W, Futai K, Karydas A et al (2012) Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep 2:789–798
Article PubMed Central CAS PubMed Google Scholar - Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646
Article PubMed Central CAS PubMed Google Scholar - Baker NE (2007) Patterning signals and proliferation in Drosophila imaginal discs. Curr Opin Genet Dev 17:287–293
Article CAS PubMed Google Scholar - Bateman JR, Lee AM, Wu CT (2006) Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173:769–777
Article PubMed Central CAS PubMed Google Scholar - Bilen J, Bonini NM (2005) Drosophila as a model for human neurodegenerative disease. Annu Rev Genet 39:153–171
Article CAS PubMed Google Scholar - Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI (2010) The nucleolus under stress. Mol Cell 40:216–227
Article PubMed Central CAS PubMed Google Scholar - Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415
CAS PubMed Google Scholar - Cleary JD, Ranum LP (2014) Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders. Curr Opin Genet Dev 26C:6–15
Article Google Scholar - DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ et al (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–256
Article PubMed Central CAS PubMed Google Scholar - Delanoue R, Legent K, Godefroy N, Flagiello D, Dutriaux A, Vaudin P et al (2004) The Drosophila wing differentiation factor vestigial–scalloped is required for cell proliferation and cell survival at the dorso-ventral boundary of the wing imaginal disc. Cell Death Differ 11:110–122
Article CAS PubMed Google Scholar - Diaz-Benjumea F, Cohen SM (1995) Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development 121:4215–4225
CAS PubMed Google Scholar - Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S et al (2013) RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80:415–428
Article PubMed Central CAS PubMed Google Scholar - Gascon E, Lynch K, Ruan H, Almeida S, Verheyden JM, Seeley WW et al (2014) Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat Med 20:1444–1451
Article PubMed Central CAS PubMed Google Scholar - Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T et al (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 126:829–844
Article PubMed Central CAS PubMed Google Scholar - Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G et al (2012) A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration–amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol 11:54–65
Article CAS PubMed Google Scholar - Guruharsha KG, Kankel MW, Artavanis-Tsakonas S (2012) The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet 13:654–666
Article PubMed Central CAS PubMed Google Scholar - Han SS, Williams LA, Eggan KC (2011) Constructing and deconstructing stem cell models of neurological disease. Neuron 70:626–644
Article CAS PubMed Google Scholar - Hetman M, Pietrzak M (2012) Emerging roles of the neuronal nucleolus. Trends Neurosci 35:305–314
Article PubMed Central CAS PubMed Google Scholar - Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S (2011) Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J Cell Biol 195:1005–1015
Article PubMed Central CAS PubMed Google Scholar - Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T et al (2014) Poly-dipeptides encoded by the C9ORF72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345:1139–1145
Article PubMed Central CAS PubMed Google Scholar - Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR et al (2013) Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci USA 110:E4530–E4539
Article PubMed Central CAS PubMed Google Scholar - Ling S-C, Polymenidou M, Cleveland DW (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79:416–438
Article PubMed Central CAS PubMed Google Scholar - Lu B, Vogel H (2009) Drosophila models of neurodegenerative diseases. Annu Rev Pathol 4:315–342
Article PubMed Central CAS PubMed Google Scholar - Mackenzie IR (2007) The neuropathology of FTD associated with ALS. Alzheimer Dis Assoc Disord 21:S44–S49
Article PubMed Google Scholar - May S, Hornburg D, Schludi MH, Arzberger T, Schwenk BM, Grässer FA et al (2014) C9orf72 FTLD/ALS-associated Gly–Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol 128:485–503
Article PubMed Central CAS PubMed Google Scholar - Mizielinska S, Lashley T, Norona FE, Clayton EL, Ridler CE, Fratta P, Isaacs AM (2013) C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol 126:845–857
Article PubMed Central CAS PubMed Google Scholar - Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A et al (2014) C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345:1192–1194
Article CAS PubMed Google Scholar - Mohan A, Goodwin M, Swanson MS (2014) RNA–protein interactions in unstable microsatellite diseases. Brain Res 1584:3–14
Article CAS PubMed Google Scholar - Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E et al (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339:1335–1338
Article CAS PubMed Google Scholar - Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR et al (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS–FTD. Neuron 72:257–268
Article PubMed Central CAS PubMed Google Scholar - Renton AE, Chiò A, Traynor B (2014) State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 17:17–23
Article PubMed Central CAS PubMed Google Scholar - Saj A, Arziman Z, Stempfle D, van Belle W, Sauder U, Horn T et al (2010) A combined ex vivo and in vivo RNAi screen for Notch regulators in Drosophila reveals an extensive Notch interaction network. Dev Cell 18:862–876
Article CAS PubMed Google Scholar - Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M et al (2013) Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med 5:208ra149
PubMed Central PubMed Google Scholar - Wang XD, Leow CC, Zha J, Tang Z, Modrusan Z, Radtke F et al (2006) Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Dev Biol 290:66–80
Article CAS PubMed Google Scholar - Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y et al (2014) Antisense proline–arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84:1213–1225
Article CAS PubMed Google Scholar - Yamakawa M, Ito D, Honda T, Kubo K, Noda M et al (2015) Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet 24:1630–1645
Article CAS PubMed Google Scholar - Yamanaka S (2007) Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell 1:39–49
Article CAS PubMed Google Scholar - Yu DX, Marchetto MC, Gage FH (2013) Therapeutic translation of iPSCs for treating neurological disease. Cell Stem Cell 12:678–688
Article CAS PubMed Google Scholar - Zhang Y-J, Jansen-West K, Xu Y-F, Gendron TF, Bieniek KF et al (2014) Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol 128:505–524
Article PubMed Central CAS PubMed Google Scholar - Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A et al (2011) Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci USA 108:260–265
Article PubMed Central CAS PubMed Google Scholar - Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O et al (2013) RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA 110:E4968–E4977
Article PubMed Central CAS PubMed Google Scholar