2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales (original) (raw)
Abstract
In March 2020, following the annual International Committee on Taxonomy of Viruses (ICTV) ratification vote on newly proposed taxa, the phylum Negarnaviricota was amended and emended. At the genus rank, 20 new genera were added, two were deleted, one was moved, and three were renamed. At the species rank, 160 species were added, four were deleted, ten were moved and renamed, and 30 species were renamed. This article presents the updated taxonomy of Negarnaviricota as now accepted by the ICTV.
Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.
Introduction
Phylum Negarnaviricota was established in 2019 by the International Committee on Taxonomy of Viruses (ICTV) for negative-sense RNA viruses that can be connected evolutionarily through their encoded RNA-directed RNA polymerase (RdRp) core domains. The phylum includes two subphyla, Haploviricotina and Polyploviricotina, for negative-sense RNA viruses that encode large (L) proteins with or without mRNA capping activity, respectively. The two subphyla include four classes (Chunqiuviricetes, Milneviricetes, Monjiviricetes, and Yunchangviricetes) and two classes (Ellioviricetes and Insthoviricetes), respectively [56, 109, 136]. The vast majority of viruses that have been assigned to phylum Negarnaviricota belong to two orders: Mononegavirales (established in 1991 [92] and amended/emended in 1995 [16], 1997 [93], 2000 [94], 2005 [95], 2011 [32], 2016 [2], 2017 [7], March 2018 [8], October 2018 [66], and 2019 [9]) and Bunyavirales (established in 2017 and amended/emended in 2018 [64, 65] and 2019 [1]).
Here we present the changes that were proposed to the entire phylum Negarnaviricota via official ICTV taxonomic proposals (TaxoProps) in 2019 and that were accepted by the ICTV in March 2020 [128]. These changes are now part of the official ICTV taxonomy.
Taxonomic changes above the phylum rank
Until recently, Negarnaviricota, included in realm Riboviria (established in 2019 [127]), was the only established phylum in the ICTV framework. In 2020, virus taxonomy was amended to include Negarnaviricota in the new riboviriad kingdom Orthornavirae as one of five sister phyla [55] (TaxoProp 2019.006G.A.v1.Riboviria).
Taxonomic changes at the subphylum rank
No new subphyla were created.
Taxonomic changes at the class rank
No new classes were created.
Taxonomic changes at the order rank
No new orders were created.
Taxonomic changes within order Goujianvirales (Haploviricotina: Yunchangviricetes)
No changes were made.
Taxonomic changes within order Jingchuvirales (Haploviricotina: Monjiviricetes)
Family Chuviridae
One new species, Taiyuan mivirus, was added to genus Mivirus for Tàiyuán leafhopper virus (TYLeV) first discovered by high-throughput sequencing (HTS) in a leafhopper (Psammotettix alienus (Dahlbom, 1850)) sampled in Tàiyuán (太原), Shānxī Province (山西省), China [129] (TaxoProp 2019.018M.A.v2.1newsp_Taiyuan_mivirus).
Taxonomic changes within order Mononegavirales (Haploviricotina: Monjiviricetes)
Family Artoviridae
The family was expanded by one new genus, Hexartovirus, including one new species, Caligid hexartovirus, for Lepeophtheirus salmonis negative-stranded RNA virus 1 (LsNSRV-1) first discovered by HTS in salmon lice (Lepeophtheirus salmonis (Krøyer, 1837)) sampled on the west coast of Norway [81]. Species Barnacle peropuvirus was moved from genus Peropuvirus into genus Hexartovirus and renamed Barnacle hexartovirus (TaxoProp 2019.021M.A.v1.1newgenus_Hexartovirus).
Family Bornaviridae
No changes were made.
Family Filoviridae
The family was expanded by one genus, Dianlovirus, including a single new species, Mengla dianlovirus, for Měnglà virus (MLAV) discovered by HTS in a Rousettus sp. bat sampled in Měnglà County (勐腊县), Yúnnán Province (云南省), China [145, 146] (TaxoProp 2019.011M.A.v1.Mengla_dianlovirus).
One new species, Bombali ebolavirus, was created in genus Ebolavirus, for Bombali virus (BOMV) first discovered by consensus PCR and confirmed by HTS in little free-tailed bats (Chaerephon pumilus (Cretzschmar, 1830–1831)) and Angolan free-tailed bats (Mops condylurus (A. Smith, 1833)) sampled in Bombali District, Northern Province, Sierra Leone [37] (TaxoProp 2019.007M.A.v2.Bombali_ebolavirus).
Family Lispiviridae
No changes were made.
Family Mymonaviridae
A new genus, Hubramonavirus, was established for two new species: Hubei hubramonavirus for Húběi rhabdo-like virus 4 (HbRLV-4) discovered by HTS in an arthropod mix collected in Húběi Province (湖北省), China [106] and Lentinula hubramonavirus for Lentinula edodes negative-strand RNA virus 1 (LeNSRV-1) first detected by HTS in commercial shiitakes (Lentinula edodes (Berk.) Pegler (1976)) sampled in Japan [60] (TaxoProp 2019.001F.A.v1.Hubramonavirus_1gen).
Family Nyamiviridae
No changes were made.
Family Paramyxoviridae
The overlooked deletion of species Bat mumps orthorubulavirus and the overlooked renaming of species Synodus paramyxovirus to Synodus synodonvirus were corrected (TaxoProp 2019.016M.A.v1.Corrections).
The family was expanded by three new genera: genus Cynoglossusvirus for the already established species Cynoglossus paramyxovirus (now renamed Cynoglossus cynoglossusvirus); genus Hoplichthysvirus for the already established species Hoplichthys paramyxovirus (now renamed Hoplichthys hoplichthysvirus); and genus Scoliodonvirus for the already established species Scoliodon paramyxovirus (now renamed Scoliodon scoliodonvirus) (TaxoProp 2019.025M.A.v2.Paramyxoviridae_3gen5sp4rensp).
Genus Aquaparamyxovirus was expanded by one species, Oncorhynchus aquaparamyxovirus, for Pacific salmon paramyxovirus (PSPV) first isolated from Chinook salmon (Oncorhynchus tshawytscha (Walbaum, 1792)) in Oregon, USA [135]. Species Salmon aquaparamyxovirus was renamed Salmo aquaparamyxovirus (TaxoProp 2019.025M.A.v2.Paramyxoviridae_3gen5sp4rensp).
Genus Jeilongvirus was expanded by one species, Miniopteran jeilongvirus, for “bat paramyxovirus isolate Bat-ParaV/B16-40” (here renamed Shaan virus [ShaV]) first isolated from a Schreibers’s long-fingered bat (Miniopterus schreibersii (Kuhl, 1817)) feces sampled in Danyang County (단양군), North Chungcheong Province (충청북도), South Korea [79] (TaxoProp 2019.025M.A.v2.Paramyxoviridae_3gen5sp4rensp).
Genus Orthoavulavirus was expanded erroneously by two species, Avian orthoavulavirus 21 and Avian orthovulavirus 21 [sic], for the same virus, “avian paramyxovirus 17” (here renamed avian paramyxovirus 21 [APMV-21]) first isolated from bird feces collected in Seosan (서산시), South Chungcheong Province (충청남도), South Korea [48] (TaxoProps 2019.014M.A.v1.Avulavirus_1newsp and 2019.025M.A.v2.Paramyxoviridae_3gen5sp4rensp).
Genus Orthorubulavirus was expanded by one species, Mammalian orthorubulavirus 6, for Alston virus (AlsV) first isolated from pteropodid bat urine sampled in Alstonville, New South Wales, Australia [49] (TaxoProp 2019.025M.A.v2.Paramyxoviridae_3gen5sp4rensp).
Genus Pararubulavirus was expanded by one species, Hervey pararubulavirus, for Hervey virus (HerV) first isolated from pteropodid bat urine sampled in Hervey Bay, Queensland, Australia [11, 53]. (TaxoProp 2019.025M.A.v2.Paramyxoviridae_3gen5sp4rensp).
Genus Respirovirus was expanded by one species, Squirrel respirovirus, for giant squirrel virus (GSqV) first isolated from a Sri Lankan giant squirrel (Ratufa macroura (Pennant, 1769)) sampled in a German zoo [35] (TaxoProp 2019.019M.A.v2.1newsp_Squirrel_respirovirus).
Family Rhabdoviridae
Genus Almendravirus was expanded by one species, Menghai almendravirus, for Menghai rhabdovirus (MRV) first isolated from Asian tiger mosquitoes (Aedes albopictus (Skuse, 1894)) collected in Měnghǎi County (勐海县) in Yúnnán Province (云南省), China [113] (TaxoProp 2019.033M.N.v1.Menghai_almendravirus_1sp).
Genus Nucleorhabdovirus was split into three genera, Alphanucleorhabdovirus, Betanucleorhabdovirus, and Gammanucleorhabdovirus (TaxoProp 2019.031M.Ac.v1.Nucleorhabdovirus_splitgen). Established species Eggplant mottled dwarf nucleorhabdovirus, Maize Iranian mosaic nucleorhabdovirus, Maize mosaic nucleorhabdovirus, Potato yellow dwarf nucleorhabdovirus, Rice yellow stunt nucleorhabdovirus, and Taro vein chlorosis nucleorhabdovirus were assigned to genus Alphanucleorhabdovirus and renamed Eggplant mottled dwarf alphanucleorhabdovirus, Maize Iranian mosaic alphanucleorhabdovirus, Maize mosaic alphanucleorhabdovirus, Potato yellow dwarf alphanucleorhabdovirus, Rice yellow stunt alphanucleorhabdovirus, and Taro vein chlorosis alphanucleorhabdovirus, respectively. Established species Datura yellow vein nucleorhabdovirus, Sonchus yellow net nucleorhabdovirus, and Sowthistle yellow vein nucleorhabdovirus were assigned to genus Betanucleorhabdovirus and renamed Datura yellow vein betanucleorhabdovirus, Sonchus yellow net betanucleorhabdovirus, and Sowthistle yellow vein betanucleorhabdovirus, respectively. Established species Maize fine streak nucleorhabdovirus was assigned to genus Gammanucleorhabdovirus and renamed Maize fine streak gammanucleorhabdovirus (TaxoProp 2019.031M.Ac.v1.Nucleorhabdovirus_splitgen).
Three new species were established in genus Alphanucleorhabdovirus:
- Morogoro maize-associated alphanucleorhabdovirus for Morogoro maize-associated virus (MMaV) first detected by HTS in maize (Zea mays L.) sampled in Morogoro, Morogoro Region, Tanzania [97];
- Physostegia chlorotic mottle alphanucleorhabdovirus for Physostegia chlorotic mottle virus (PhCMoV) first isolated from lionhearts (Physostegia sp.) in Austria [76]; and
- Wheat yellow striate alphanucleorhabdovirus for wheat yellow striate virus (WYSV) first isolated from common wheat (Triticum aestivum L.) sampled in Hánchéng (韩城), Shǎnxī/Shaanxi Province (陕西省), China [61] (TaxoProp 2019.031M.Ac.v1.Nucleorhabdovirus_splitgen).
Three new species were established in genus Betanucleorhabdovirus:
- Alfalfa betanucleorhabdovirus for alfalfa-associated nucleorhabdovirus (AaNV) first discovered by HTS in alfalfa (Medicago sativa L.) sampled in Stadl-Paura, Upper Austria (Oberösterreich), Austria [36];
- Blackcurrant betanucleorhabdovirus for blackcurrant-associated rhabdovirus (BCaRV) first discovered by HTS in blackcurrant (Ribes nigrum L.) sampled in Russia [138]; and
- Trefoil betanucleorhabdovirus for birds-foot trefoil-associated virus (BFTV) first discovered by HTS in Bird’s-foot trefoil (Lotus corniculatus L.) sampled in the Qínlǐng Mountains (秦岭山), Shǎnxī/Shaanxi Province (陕西省), China [26, 131] (TaxoProp 2019.031M.Ac.v1.Nucleorhabdovirus_splitgen).
One new genus, Arurhavirus, was established to include four species:
- Aruac arurhavirus for Aruac virus (ARUV) first isolated from mosquitoes (Trichoprosopon theobaldi Lane and Cerqueira, 1942) collected in Melaju Forest, Trinidad, Trinidad and Tobago [110, 126];
- Inhangapi arurhavirus for Inhangapi virus (INHV) first isolated from sandflies (Lutzomyia flaviscutellata (Mangabeira, 1942)) collected in Catu Forest, Belém, Pará State, Brazil [3, 126];
- Santabarbara arurhavirus for Santa Barbara virus (SBAV) first in mice sampled in Santa Bárbara do Pará, Pará State, Brazil [unpublished]; and
- Xiburema arurhavirus for Xiburema virus (XIBV) first isolated from mosquitoes (Sabethes intermedius (Lutz, 1904)) sampled in Sena Madureira, Acre State, Brazil [51, 132] (2019.006M.A.v1.Arurhavirus_1gen4sp).
One new genus, Barhavirus, was established to include two new species:
- Bahia barhavirus for Bahia Grande virus (BGV) first isolated from mosquitoes (Aedes, Culex, Anopheles, Psorophora spp.) collected in Texas, Louisiana, New Mexico, and North Dakota, USA [52, 126] and also for Harlingen virus (HARV) isolated from salt marsh mosquitoes (Culex salinarius Coquillett, 1904) sampled in Harlingen, Texas, USA [126].
- Muir barhavirus for Muir Springs virus (MSV) first isolated from mosquitoes (Aedes sp.) collected in Fort Morgan, Colorado, USA [52, 126] (TaxoProp 2019.012M.A.v1.Rhabdoviridae_5gen8sp1reasp).
One new genus, Lostrhavirus, was established to include new species Lonestar zarhavirus [sic] for lone star tick rhabdovirus (LSTRV) first detected by HTS in lone star ticks (Amblyomma americanum (Linnaeus, 1758)) collected in the USA [unpublished] (TaxoProp 2019.012M.A.v1.Rhabdoviridae_5gen8sp1reasp).
One new genus, Mousrhavirus, was established to include the previously established species Moussa virus (now renamed Moussa mousrhavirus) (TaxoProp 2019.012M.A.v1.Rhabdoviridae_5gen8sp1reasp).
One new genus, Ohlsrhavirus, was established to include five new species:
- Culex ohlsrhavirus for Culex rhabdo-like virus (CRLV) first discovered by HTS in southern house mosquitoes (Culex quinquefasciatus Say, 1823) collected near Perth, Western Australia, Australia [107];
- Northcreek ohlsrhavirus for North Creek virus (NORCV) first discovered by HTS in mosquitoes (Culex sitiens Wiedemann, 1828) collected in Ballina, New South Wales, Australia [23];
- Ohlsdorf ohlsrhavirus for Ohlsdorf virus (OHLDV) first discovered by HTS in mosquitoes (Ochlerotatus cantans (Meigen, 1818)) collected in Hamburg, Germany [105];
- Riverside ohlsrhavirus for riverside virus (RISV) first discovered by HTS in mosquitoes (Ochlerotatus sp.) collected in Gemenc, Gyékényes, and Drávaszabolcs, Hungary [98]; and
- Tongilchon ohlsrhavirus for Tongilchon virus 1 (TCHV-1) first detected in mosquitoes (Culex bitaeniorhynchus Giles, 1901) collected in Tongil-chon (통일촌), Gyeonggi Province (경기도), South Korea [39] (TaxoProp 2019.032M.N.v1.Ohlsrhavirus_1gen5sp).
One new genus, Sawgrhavirus, was established to include four new species:
- Connecticut sawgrhavirus for Connecticut virus (CNTV) first isolated from ticks (Ixodes dentatus Marx, 1899) taken from an eastern cottontail (Sylvilagus floridanus (J. A. Allen, 1890)) captured in Lyme, Connecticut, USA [67, 126];
- Island sawgrhavirus for Long Island tick rhabdovirus (LITRV) first detected by HTS in lone star ticks (Amblyomma americanum (Linnaeus, 1758)) collected on Long Island, New York, USA [119];
- Minto sawgrhavirus for New Minto virus (NMV) first isolated from rabbit ticks (Haemaphysalis leporispalustris Packard, 1869) sampled in New Minto, Alaska, USA [99, 126]; and
- Sawgrass sawgrhavirus for Sawgrass virus (SAWV) isolated from American dog ticks (Dermacentor variabilis (Say, 1821)) sampled at Sawgrass Lake, Tampa Bay, Florida, USA [102, 126] (TaxoProp 2019.012M.A.v1.Rhabdoviridae_5gen8sp1reasp).
One new genus, Sunrhavirus, was established to accommodate six novel species:
- Garba sunrhavirus for Garba virus (GARV) first isolated from a malachite kingfisher (Corythornis cristatus (Pallas, 1764)) trapped in Bangui, Central African Republic [51, 126];
- Harrison sunrhavirus for Harrison Dam virus (HARDV) first isolated from common banded mosquitoes (Culex annulirostris Skuse, 1889) collected at Beatrice Hill, Northern Territory, Australia [71];
- Kwatta sunrhavirus for Kwatta virus (KWAV) first isolated from mosquitoes (Culex sp.) collected near Paramaribo, Suriname [25, 126];
- Oakvale sunrhavirus for Oak Vale virus (OVV) first isolated from mosquitoes (Culex edwardsi Barraud, 1923) sampled in Peachester, Queensland, Australia [77, 96];
- Sunguru sunrhavirus for Sunguru virus (SUNV) first isolated from a domestic chicken (Gallus gallus domesticus (Linnaeus, 1758)) in Arua District, Northern Region, Uganda [58]; and
- Walkabout sunrhavirus for Walkabout Creek virus (WACV) first isolated from biting midges (Culicoides austropalpalis Lee and Reye, 1955) collected near Samford, Queensland, Australia [71] (2019.004M.A.v2.Sunrhavirus).
One new genus, Zarhavirus, was created for one new species, Zahedan zarhavirus, for Zahedan rhabdovirus (ZARV) first isolated from ticks (Hyalomma anatolicum anatolicum (Koch, 1844)) collected in Zâhedân  , Sistan and Baluchestan Province
, Sistan and Baluchestan Province  , Iran [28] (TaxoProp 2019.012M.A.v1.Rhabdoviridae_5gen8sp1reasp).
, Iran [28] (TaxoProp 2019.012M.A.v1.Rhabdoviridae_5gen8sp1reasp).
One new species, Taiwan bat lyssavirus, was added to genus Lyssavirus for Taiwan bat lyssavirus (TWBLV) first isolated from a Japanese pipistrelle (Pipistrellus abramus (Temminck, 1838)) sampled in Taiwan [45] (TaxoProp 2019.001M.A.v1.Lyssavirus).
Genus Cytorhabdovirus was expanded by 12 species:
- Cabbage cytorhabdovirus for cabbage cytorhabdovirus 1 (CCyV-1) first discovered by HTS in cabbage (Brassica oleracea L.) sampled in the UK [89];
- Maize-associated cytorhabdovirus for maize-associated cytorhabdovirus (MaCV) first discovered by HTS in maize (Zea mays L.) collected in Lima, Peru [133];
- Maize yellow striate cytorhabdovirus for maize yellow striate virus (MYSV) first discovered by HTS in maize (Zea mays L.) and common wheat (Triticum aestivum L.) collected in Sinsacate, Córdoba Province, Argentina [70];
- Papaya cytorhabdovirus for papaya virus E (PpVE) first discovered by HTS in papaya (Carica papaya L.) sampled in Los Ríos Province, Ecuador [73];
- Persimmon cytorhabdovirus for persimmon virus A (PeVA) first discovered by HTS in Japanese persimmon (Diospyros kaki L.f.) sampled in Japan [47];
- Raspberry vein chlorosis cytorhabdovirus for raspberry vein chlorosis virus (RVCV) first discovered by HTS in red raspberries (Rubus idaeus L.) sampled in Dundee, Scotland, UK [50];
- Rice stripe mosaic cytorhabdovirus for rice stripe mosaic virus (RSMV) first discovered by HTS in rice (Oryza sativa L.) sampled in Luódìng (罗定), Guǎngdōng Province (广东省), China [147];
- Tomato yellow mottle-associated cytorhabdovirus for tomato yellow mottle-associated virus (TYMaV) first discovered by HTS in tomato (Solanum lycopersicum L.) sampled in Chóngqìng (重庆), China [141];
- Wuhan 4 insect cytorhabdovirus for Wuhan insect virus 4 (WuIV-4) first discovered by HTS in mealy plum aphids (Hyalopterus pruni (Geoffroy, 1762)) sampled in Wǔhàn (武汉), Húběi Province (湖北省), China [59];
- Wuhan 5 insect cytorhabdovirus for Wuhan insect virus 5 (WuIV-5) first discovered by HTS in mealy plum aphids (Hyalopterus pruni (Geoffroy, 1762)) sampled in Wǔhàn (武汉), Húběi Province (湖北省), China [59];
- Wuhan 6 insect cytorhabdovirus for Wuhan insect virus 6 (WuIV-6) first discovered by HTS in mealy plum aphids (Hyalopterus pruni (Geoffroy, 1762)) sampled in Wǔhàn (武汉), Húběi Province (湖北省), China [59]; and
- Yerba mate chlorosis-associated cytorhabdovirus for yerba mate chlorosis-associated virus (YmCaV) [12] first discovered by HTS in yerba mate (Ilex paraguariensis A. St.-Hil.) sampled in Cerro Azul, Misiones Province, Argentina (TaxoProps 2019.002M.A.v3.Cytorhabdovirus and 2019.030M.A.v1.Cytorhabovirus_12newsp).
One new species, Holmes hapavirus, was added to genus Hapavirus for Holmes Jungle virus (HOJV) first isolated from common banded mosquitoes (Culex annulirostris Skuse, 1889) collected near Darwin, Northern Territory, Australia [38] (2019.003M.A.v3.Hapavirus).
Three new species were added to genus Sripuvirus:
- Charleville sripuvirus for Charleville virus (CHVV) first isolated from sandflies (Phlebotomus sp.) collected in Charleville, Queensland, Australia [29, 123];
- Cuiaba sripuvirus for Cuiaba virus (CUIV) isolated from a cane toad (Rhinella marina (Linnaeus, 1758)) captured in Pará State, Brazil [51, 123]; and
- Hainan sripuvirus for Hainan black-spectacled toad rhabdovirus (HnBSTRV) first detected by HTS in an Asian common toad (Duttaphrynus melanostictus (Schneider, 1799)) sampled in Hǎinán Province (海南省), China [108] (TaxoProp 2019.013M.A.v1.Sripuvirus_3newsp).
Taxonomic changes within order Muvirales (Haploviricotina: Chunqiuviricetes)
No changes were made.
Taxonomic changes within order Serpentovirales (Haploviricotina: Milneviricetes)
No changes were made.
Taxonomic changes within order Articulavirales (Polyploviricotina: Insthoviricetes)
No changes were made.
Taxonomic changes within order Bunyavirales (Polyploviricotina: Ellioviricetes)
Family Arenaviridae
Genus Hartmanivirus was expanded by three species: Muikkunen hartmanivirus for Dante Muikkunen virus 1 (DaMV-1), Schoolhouse hartmanivirus for old schoolhouse viruses 1 and 2 (OScV-1/2), and Zurich hartmanivirus for veterinary pathology Zurich viruses 1 and 2 (VPZV-1/2), all first detected by HTS in captive boid snakes [44] (TaxoProp 2019.008M.A.v2.Hartmanivirus_3new sp).
Genus Mammarenavirus was expanded by four species:
- Alxa mammarenavirus for RtDs-AreV/IM2014 virus (here renamed Alxa virus [ALXV]) (TaxoProp 2019.020M.A.v2.1newsp_Alxa_mammarenavirus) first discovered by HTS in a Northern three-toed jerboa (Dipus sagitta (Pallas, 1773)) sampled in Alxa Left Banner (阿拉善左旗), Inner Mongolia Autonomous Region (内蒙古自治区), China [139, 140];
- Chevrier mammarenavirus for Lìjiāng virus (LIJV) first discovered by HTS in a Chevrier’s field mouse (Apodemus chevrieri (Milne-Edwards, 1868)) sampled around Lìjiāng (丽江), Yúnnán Province (云南省), China [unpublished] (TaxoProp 2019.009M.A.v2.Mammarenavirus_sp_LIJV);
- Planalto mammarenavirus for Aporé virus (APOV) first discovered by HTS in a Mato Grosso colilargo (Oligoryzomys mattogrossae (J. A. Allen, 1916)) sampled in Cassilândia, Mato Grosso do Sul State, Brazil (TaxoProp 2019.010M.A.v1.Mammarenavirus_sp_APOV) [34]; and
- Xapuri mammarenavirus for Xapuri virus (XAPV) first discovered by HTS in a Musser’s neacomys (Neacomys musseri Patton, da Silva, and Malcolm, 2000) sampled in Xapuri, Acre State, Brazil [33] (TaxoProp 2019.005M.A.v1.Mammarenavirus_sp_XAPV).
Family Fimoviridae
Genus Emaravirus was expanded by two species: Blackberry leaf mottle associated emaravirus for blackberry leaf mottle-associated virus (BLMaV) first discovered in blackberries (Rubus spp.) collected in various US states (TaxoProp 2019.010P.A.v1.Emaravirus_1sp) [41] and Pistacia emaravirus B for pistacia virus B (PiVB) discovered by HTS in pistachios (Pistacia vera L.) sampled in Turkey [18] (TaxoProp 2019.011P.A.v1.Emaravirus_1sp).
Family Hantaviridae
Genus Loanvirus was expanded by one species, Brno loanvirus, for Brno virus (BRNV) first discovered by HTS in a noctule (Nyctalus noctula (Schreber, 1774)) sampled in Brno, South Moravia Region (Jihomoravský kraj), Czech Republic [112] (TaxoProp 2019.017M.A.v3.1newsp_Brno_virus).
Family Peribunyaviridae
Genus Pacuvirus was expanded by two species: new species Caimito pacuvirus for Caimito virus (CAIV) first isolated from sandflies (Nyssomyia ylephiletor (Fairchild and Hertig, 1952)) sampled in El Aguacate, Panamá Province, Panama [46, 116] (TaxoProp 2019.022M.A.v2.2sp_Pacuvirus) and Chilibre pacuvirus (the former Chilibre phlebovirus, renamed and moved from genus Phlebovirus) (TaxoProps 2019.022M.A.v2.2sp_Pacuvirus and 2019.026M.A.v1.Phenuiviridae_4gen79sp).
Family Phasmaviridae
The previously established genus Inshuvirus and its included species Insect inshuvirus were both abolished due to insufficient member virus information (TaxoProp 2019.028M.A.v2.Phasmaviridae_1newsp_abol1gen3sp).
New species Anopheles orthophasmavirus was included in genus Orthophasmavirus for Anopheles triannulatus orthophasmavirus (AtOPV) first discovered by HTS in mosquitoes (Anopheles triannulatus (Neiva and Pinto, 1922)) sampled in Santa Bárbara Farm, Amapá State, Brazil [103]. Two species, Nome phantom orthophasmavirus and Seattle orthophasmavirus, were abolished (TaxoProp 2019.028M.A.v2.Phasmaviridae_1newsp_abol1gen3sp).
Family Phenuiviridae
The previously unassigned genus Coguvirus was included in family Phenuiviridae (TaxoProp 2019.026M.A.v1.Phenuiviridae_4gen79sp). One new species, Coguvirus eburi, was created in the genus for citrus virus A (CiVA) first discovered by HTS in a sweet orange tree in Italy [78] (2019.004P.A.v1.Coguvirus_1sp).
Genus Banyangvirus and included species Huaiyangshan banyangvirus, Guertu banyangvirus, and Heartland banyangvirus were renamed Bandavirus, Dabie bandavirus, Guertu bandavirus, and Heartland bandavirus, respectively (TaxoProps 2019.015M.A.v1.Bandavirus and 2019.026M.A.v1.Phenuiviridae_4gen79sp). Four new bandavirus species were added to the genus:
- Bhanja bandavirus for Bhanja virus (BHAV) first isolated from flat-inner-spurred haemaphysalids (Haemaphysalis intermedia Warburton and Nuttall, 1909) sampled in Orissa State, India [27, 104];
- Hunter Island bandavirus for Hunter Island virus (HUIV) first isolated from ticks (Ixodes eudyptidis Maskell, 1885) sampled on Albatross Island, Tasmania, Australia [130];
- Kismaayo bandavirus for Kismaayo virus (KISV; name corrected from the previously circulating “Kismayo virus” and “Kisemayo virus”) first isolated from yellow back ticks (Rhipicephalus pulchellus (Gerstäcker, 1873)) sampled in Kismaayo, Lower Juba (Jubbada Hoose) Region, Somalia [149]; and
- Lone Star bandavirus [sic] for lone star virus (LSV) first isolated from lone star ticks (Amblyomma americanum (Linnaeus, 1758)) sampled in Kentucky, USA [54, 114] (TaxoProp 2019.026M.A.v1.Phenuiviridae_4gen79sp).
Genus Entovirus was created for one new species, Entoleuca entovirus, for Entoleuca phenui-like virus 1 (EnPLV-1) first discovered by HTS in Entoleuca sp. fungi sampled in Málaga Province, Spain [124] (TaxoProp 2019.026M.A.v1.Phenuiviridae_4gen79sp).
Genus Kabutovirus and included species Kabuto mountain kabutovirus and Huangpi kabutovirus were renamed Uukuvirus, Kabuto mountain uukuvirus, and Huangpi uukuvirus, respectively. The established species Uukuniemi phlebovirus was moved into genus Uukuvirus and renamed Uukuniemi uukuvirus. 14 new species were established in genus Uukuvirus:
- American dog uukuvirus for American dog tick virus (ADAV) first detected by HTS in American dog ticks (Dermacentor variabilis (Say, 1821)) sampled in Heckscher State Park, New York, USA [120];
- Dabieshan uukuvirus for Dàbiéshān tick virus (DbsTV) first discovered by HTS in Asian longhorned ticks (Haemaphysalis longicornis Neumann, 1901) collected in the Dàbié Mountains (大別山), China [59];
- Grand Arbaud uukuvirus for Grand Arbaud virus (GAV) first isolated from ticks (Argas reflexus (Fabricius, 1794)) sampled in Bouches-du-Rhône Department, France [40, 85];
- Kaisodi uukuvirus for Kaisodi virus (KASDV) first isolated from hard-bodied ticks (Haemaphysalis spinigera Neumann, 1897) sampled in Mysore State, India [14, 88, 144];
- Lihan uukuvirus for Lǐhán tick virus (LITV) first discovered by HTS in Asian blue ticks (Rhipicephalus microplus (Canestrini, 1888)) sampled in Lǐhán (李韩), Húběi Province (湖北省), China [59];
- Murre uukuvirus for murre virus (MURV) first isolated from common murres (Uria aalge (Pontoppidan, 1763)) sampled in Alaska, USA [85];
- Pacific coast uukuvirus for Pacific coast tick virus (PACTV) first discovered by HTS in Pacific coast ticks (Dermacentor occidentalis Marx, 1892) sampled in Mendocino County, California, USA [17];
- Precarious Point uukuvirus for Precarious Point virus (PPV) first isolated from seabird ticks (Ixodes uriae White, 1852) sampled on Macquarie Island, Tasmania, Australia [85, 111];
- Rukutama uukuvirus for Rukutama virus (RUKV) first isolated from seabird ticks (Ixodes uriae White, 1852) sampled on Tûlenij/Tyuleny Island (Ocтpoв Tюлeний), Sakhalin Oblast (Caxaлинcкaя oблacть), Russia [63, 150];
- Schmidt uukuvirus for EgAn 1825-61 virus (here renamed Nile warbler virus [NIWV]) first isolated from a willow warbler (Phylloscopus trochilus (Linnaeus, 1758)) sampled in Nile Delta, Egypt [85];
- Silverwater uukuvirus for Silverwater virus (SILV) first isolated from rabbit ticks (Haemaphysalis leporispalustris Packard, 1869) sampled near Powassan, Ontario, Canada [69, 72];
- Tacheng uukuvirus for Tǎchéng tick virus 2 (TcTV-2) first discovered by HTS in ticks (Dermacentor marginatus Sulzer, 1776) sampled in China [59];
- Yongjia uukuvirus for Yǒngjiā tick virus 1 (YjTV-1) first discovered by HTS in East Asian mountain haemaphysalids (Haemaphysalis hystricis Supino, 1897) in China [59]; and
- Zaliv Terpeniya uukuvirus for Zaliv Terpeniya virus (ZTV) first isolated from seabird ticks (Ixodes uriae White, 1852) sampled on Tyuleny Island (Tюлeний ocтpoв) in the Gulf of Patience (Зaлив Tepпeния), Sakhalin Oblast (Caxaлинcкaя oблacть) and Commander Islands (Кoмaндopcкиe ocтpoвa), Kamchatka Krai (Кaмчaтcкий кpaй), RSFSR, USSR [62, 151] (TaxoProp 2019.026M.A.v1.Phenuiviridae_4gen79sp).
Genus Ixovirus was established for the three new species:
- Blackleg ixovirus for blacklegged tick phlebovirus 1, here renamed blacklegged tick virus 1 (BLTV-1), first discovered by HTS in deer ticks (Ixodes scapularis Say, 1821) sampled in Heckscher State Park, New York, USA [120];
- Norway ixovirus for Norway phlebovirus 1, here renamed Fairhair virus (FHAV), first discovered by HTS in castor bean ticks (Ixodes ricinus (Linnaeus, 1758)) sampled in Norway [91]; and
- Scapularis ixovirus for blacklegged tick phlebovirus 3, here renamed blacklegged tick virus 3 (BLTV-3), first discovered by HTS in deer ticks (Ixodes scapularis Say, 1821) sampled in Heckscher State Park, New York, USA [120] (TaxoProp 2019.026M.A.v1.Phenuiviridae_4gen79sp).
Genus Lentinuvirus was created for one new species, Lentinula lentinuvirus, for Lentinula edodes negative-strand RNA virus 2 (LeNSRV-2) first discovered by HTS in shiitakes (Lentinula edodes (Berk.) Pegler (1976)) sampled in Japan [60] (TaxoProp 2019.026M.A.v1.Phenuiviridae_4gen79sp).
In genus Phlebovirus, established species Sandfly fever Naples phlebovirus was renamed Naples phlebovirus. The genus was expanded by 53 new species (TaxoProp 2019.026M.A.v1.Phenuiviridae_4gen79sp):
- Adana phlebovirus for Adana virus (ADAV) first isolated from Phlebotomus spp. sandflies sampled in Adana, Adana Province (Adana ili), Turkey [4];
- Aguacate phlebovirus for Aguacate virus (AGUV) first isolated from Lutzomyia spp. sandflies sampled in El Aguacate, Panamá Province, Panama [82, 116];
- Alcube phlebovirus for Alcube virus (ACBV) first isolated from sandflies (Phlebotomus perniciosus Newstead, 1911) sampled around Arrábida, Portugal [10];
- Alenquer phlebovirus for Alenquer virus (ALEV) first isolated from a human in Ramal das Pias, Alenquer, Pará State, Brazil [83, 122];
- Ambe phlebovirus for Ambe virus (ABEV) first isolated from psychodid sandflies sampled near Altamira, Pará State, Brazil [80, 118];
- Anhanga phlebovirus for Anhangá virus (ANHV) first isolated from a Linnaeus’s two-toed sloth (Choloepus didactylus (Linnaeus, 1758)) sampled in Castanhal Forest, Pará State, Brazil [80];
- Arumowot phlebovirus for Arumowot virus (AMTV) first isolated from mosquitoes (Culex antennatus (Becker, 1903)) sampled in Sudan [13, 84];
- _Buenaventura phleboviru_s for Buenaventura virus (BUEV) first isolated in 1984 from Lutzomyia sp. sandflies sampled in Rio Raposo, Valle del Cauca Department, Colombia [87, 116];
- Cacao phlebovirus for Cacao virus (CACV) first isolated from sandflies (Nyssomyia trapidoi (Fairchild and Hertig, 1952)) sampled in El Aguacate, Panamá Province, Panama [87, 116];
- Campana phlebovirus for Campana virus (CMAV) first isolated from phlebotomine sandflies sampled in El Aguacate, Panamá Province, Panama [87];
- Chagres phlebovirus for Chagres virus (CHGV) first isolated from a human sampled at Fort Sherman, Canal Zone/Cólon Province, Panama [90];
- Cocle phlebovirus for Coclé virus (CCLV) first isolated from a human sampled in Penonomé, Coclé Province, Panama [87];
- Dashli phlebovirus for Dāshlī virus (DASV) first isolated from Sergentomyia sp. sandflies sampled in Dāshlīborun
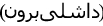 , Golestān Province
, Golestān Province 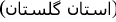 , Iran [6];
, Iran [6]; - Durania phlebovirus for Durania virus (DRNV) first isolated from sandflies sampled in 1986 near Durania, North Santander Department, Colombia [82, 118];
- Echarate phlebovirus for Echarate virus (ECHV) first isolated from a human sampled in Cusco, Peru [83];
- Gabek phlebovirus for Gabek Forest virus (GFV) first isolated from a northeast African spiny mouse (Acomys cahirinus (É. Geoffroy, 1803)) sampled in Gabek Forest, near Paloich, Sudan [86];
- Gordil phlebovirus for Gordil virus (GORV) first isolated from a typical lemniscomys (Lemniscomys striatus (Linnaeus, 1758)) sampled in Gordil, Vakaga Prefecture, Central African Republic [86];
- Icoaraci phlebovirus for Icoaraci virus (ICOV) first isolated from from an unidentified forest rat sampled in Belém, Pará State, Brazil [19, 142];
- Itaituba phlebovirus for Itaituba virus (ITAV) first isolated from a common opossum (Didelphis marsupialis Linnaeus, 1758) trapped at the Tapacurazinho stream, Itaituba, Pará State, Brazil [83, 122];
- Itaporanga phlebovirus for Itaporanga virus (ITPV) first isolated from a sentinel Swiss mouse collected in Itaporanga, São Paulo State, Brazil [46, 121];
- Ixcanal phlebovirus for Ixcanal virus (IXCV) first isolated from Lutzomyia sp. sandflies sampled in Aldea Ixcanal and Aldea Puerta, El Progreso Departmesp. sandflies innt, Guatemala [82, 118];
- Karimabad phlebovirus for Karimabad virus (KARV) first isolated from Phlebotomus sp. sandflies in Karīmābād
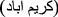 , Khūzestān Province
, Khūzestān Province 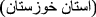 , Iran [86];
, Iran [86]; - La Gloria phlebovirus for La Gloria virus (LAGV) first discovered by HTS in phlebotomine sandflies sampled near La Gloria village, Panama Canal area, central Panama [68];
- Lara phlebovirus for GGP-2011a virus (here renamed Rio Claro virus [RICV]) first isolated from a sentinel hamster sampled in Venezuela [unpublished];
- Leticia phlebovirus for Leticia virus (LETV) first isolated from sandflies sampled in Leticia, Amazonas Department, Colombia [87];
- Maldonado phlebovirus for Maldonado virus (MLOV) first isolated from a human sampled in Puerto Maldonado, Madre de Dios Region, Peru [83];
- Massilia phlebovirus for Massilia virus (MASV) first isolated from sandflies (Phlebotomus perniciosus Newstead, 1911) sampled in Marseille and Nice, Provence-Alpes-Côte d’Azur, France [20, 86];
- Medjerda phlebovirus for Medjerda Valley virus (MVV) first isolated from phlebotomine sandflies sampled at an archaeological site in Bizerte Governorate, Tunisia [15];
- Mona Grita phlebovirus for Mona Grita virus (MOGV) first discovered by HTS in sandflies (Nyssomyia trapidoi (Fairchild and Hertig, 1952)) sampled on Isla Mona Grita, Panama Canal, central Panama [68];
- Munguba phlebovirus for Munguba virus (MUNV) first isolated from sandflies (Nyssomyia umbratilis (Ward and Fraiha, 1977)) sampled in Monte Dourado, Pará State, Brazil [80, 122];
- Nique phlebovirus for Nique virus (NIQV) first isolated from sandflies (Lutzomyia panamensis (Shannon, 1926)) sampled in Cerro Nique, Darién Province, Panama [83, 117];
- Ntepes phlebovirus for Ntepes virus (NTPV) first isolated from Sergentomyia sp. sandflies sampled near Ntepes village, Marigat District, Baringo County, Kenya [115];
- Odrenisrou phlebovirus for Odrénisrou virus (ODRV) first isolated from mosquitoes (Culex albiventris Edwards, 1922) collected in the forest of Taï National Park, Côte d’Ivoire [84];
- Oriximina phlebovirus for Oriximiná virus (ORXV) first isolated from Lutzomyia sp. sandflies sampled in Saracazinho, Pará State, Brazil [83, 122];
- Pena Blanca phlebovirus for Peña Blanca virus (PEBV) first discovered by HTS in sandflies sampled on Peña Blanca peninsula, Panama Canal, central Panama [68];
- Punique phlebovirus for Punique virus (PUNV) first isolated from sandflies (Phlebotomus perniciosus Newstead, 1911 and Phlebotomus longicuspis Nitzulescu, 1930) sampled in Tunis, Tunisia [86];
- Rio Grande phlebovirus for Rio Grande virus (RGV) first isolated from a Southern Plains woodrat (Neotoma micropus Baird, 1855) sampled in Texas, USA [46];
- Saint Floris phlebovirus for Saint-Floris virus (SAFV) first isolated from a gerbil sampled in Gordil, Vakaga Prefecture, Central African Republic [86];
- Salanga phlebovirus for Salanga virus (SLGV) first isolated from a Hinde’s aethomys (Aethomys hindei (Thomas, 1902)) collected in Salanga, Ombella-M’Poko Prefecture, Central African Republic [51, 148];
- Salobo phlabovirus [sic] for Salobo virus (SLBOV) first isolated from a Guyenne spiny-rat (Proechimys guyannensis (E. Geoffroy, 1803)) in Pará State, Brazil [142];
- Sicilian phlebovirus for sandfly fever Sicilian virus (SFSV) first isolated from a human sampled in Palermo Province, Sicily Region, Italy [101, 137];
- Tapara phlebovirus for Tapará virus (TPRV) first isolated from phlebotomine sandflies in Altamira, Pará State, Brazil [80];
- Tehran phlebovirus for Tehran virus (THEV) first isolated from sandflies (Phlebotomus papatasi (Scopoli, 1786)) sampled in Tehran, Iran [86];
- Tico phebovirus [sic] for Tico virus (TICV) discovered by HTS in sandflies sampled in Panama Canal area, central Panama [68];
- Toros phlebovirus for Toros virus (TORV) first discovered by HTS in sandflies sampled in Damyeri, Adana Province (Adana ili), Turkey [5];
- Toscana phlebovirus for Toscana virus (TOSV) first isolated from sandflies (Phlebotomus perniciosus Newstead, 1911) in Toscany, Italy [86, 125];
- Tres Almendras phlebovirus for Tres Almendras virus (TRAV) first discovered by HTS in sandflies (Psychodopygus panamensis (Shannon, 1926)) sampled on Tres Almendras Islands, Panama Canal area, central Panama [68];
- Turuna phlebovirus for Turuna virus (TUAV) first isolated from Lutzomyia sp. sandflies sampled in Cachoeira Porteira, Pará State, Brazil [83, 122];
- Uriurana phlebovirus for Uriurana virus (URIV) first isolated from phlebotomine sandflies in Tucuruí, Pará State, Brazil [80];
- Urucuri phlebovirus for Urucuri virus (URUV) first isolated from a Guyenne spiny-rat (Proechimys guyannensis (E. Geoffroy, 1803)) in Utinga Forest, Belém, Pará State, Brazil [80, 122];
- Viola phlebovirus for viola virus (VIOV) first discovered by HTS in sandflies (Lutzomyia longipalpis (Lutz and Neiva, 1912)) sampled in Pirizal, Mato Grosso State, Brazil [24]; and
- Zerdali phlebovirus for Zerdali virus (ZERV) first discovered by HTS in sandflies sampled in Zerdali, Adana Province (Adana ili), Turkey [5].
Genus Rubodvirus was created for the two new species Apple rubodvirus 1 and 2 to accommodate apple rubbery wood viruses 1 and 2 (ARWV-1/2), respectively, first discovered using HTS in apple trees (Malus sp.) sampled in Germany and USA [100] (TaxoProp 2019.026M.A.v1.Phenuiviridae_4gen79sp).
One new species, Melon tenuivirus, was added to genus Tenuivirus for melon chlorotic spot virus (MeCSV) first isolated from muskmelon (Cucumis melo L.) sampled in Provence-Alpes-Côte d’Azur Region, France [57].
Genus Wubeivirus was abolished and its two species, Fly wubeivirus and Dipteran wubeivirus, were moved into genus Phasivirus and renamed Fly phasivirus and Dipteran phasivirus, respectively (TaxoProp 2019.026M.A.v1.Phenuiviridae_4gen79sp).
Family Tospoviridae
The overlooked adjustment of 12 tospovirid species names to correct non-Latinized binomials was implemented (TaxoProp 2019.016M.A.v1.Corrections).
Eight new species were created in genus Orthotospovirus:
- Alstroemeria necrotic streak orthotospovirus for Alstroemeria necrotic streak virus (ANSV) first isolated from ornamental crops (Alstroemeria sp.) sampled in Colombia [42];
- Alstroemeria yellow spot orthotospovirus for Alstroemeria yellow spot virus (AYSV) first isolated from ornamental crops (Alstroemeria sp.) imported to and sampled in the Netherlands [43];
- Groundnut chlorotic fan spot orthotospovirus for groundnut chlorotic fan-spot virus (GCFSV) first isolated from peanut (Arachis hypogaea L.) sampled in Taiwan [21];
- Hippeastrum chlorotic ringspot orthotospovirus for Hippeastrum chlorotic spot virus (HCRV) first isolated from amaryllis (Hippeastrum sp.) and spider lily (Hymenocallis littoralis (Jacq.) Salisb.) sampled in southwestern China [31, 143];
- Mulberry vein banding associated orthotospovirus for mulberry vein banding-associated virus (MVBaV) discovered first by HTS in mulberry (Morus alba L.) sampled in Guǎngxī Zhuàng Autonomous Region (广西壮族自治区), China in 2011 [74, 75];
- Pepper chlorotic spot orthotospovirus for pepper chlorotic spot virus (PCSV) first isolated from sweet pepper (Capsicum annuum L.) in Taiwan [22];
- Tomato yellow ring orthotospovirus for tomato yellow ring virus (TYRV) first isolated from tomato (Solanum lycopersicum L.) in Iran [134]; and
- Tomato zonate spot orthotospovirus for tomato zonate spot virus (TZSV) first isolated from tomato (Solanum lycopersicum L.) and chili pepper (Capsicum annuum L.) sampled in Yúnnán Province (云南省), China [30] (TaxoProp 2019.006P.A.v1.Orthotospovirus_8sp).
SUMMARY
A summary of the current, ICTV-accepted taxonomy of the phylum Negarnaviricota is presented in Table 1 (Goujianvirales), Table 2 (Jingchuvirales), Table 3 (Mononegavirales), Table 4 (Muvirales), Table 5 (Serpentovirales), Table 6 (Articulavirales), and Table 7 (Bunyavirales).
Table 1 ICTV-accepted taxonomy of the order Goujianvirales (Negarnaviricota: Haploviricotina: Yunchangviricetes) as of March 2020
Table 2 ICTV-accepted taxonomy of the order Jingchuvirales (Negarnaviricota: Haploviricotina: Monjiviricetes) as of March 2020
Table 3 ICTV-accepted taxonomy of the order Mononegavirales (Negarnaviricota: Haploviricotina: Monjiviricetes) as of March 2020
Table 4 ICTV-accepted taxonomy of the order Muvirales (Negarnaviricota: Haploviricotina: Chunqiuviricetes) as of March 2020
Table 5 ICTV-accepted taxonomy of the order Serpentovirales (Negarnaviricota: Haploviricotina: Milneviricetes) as of March 2020
Table 6 ICTV-accepted taxonomy of the order Articulavirales (Negarnaviricota: Polyploviricotina: Insthoviricetes) as of March 2020
Table 7 ICTV-accepted taxonomy of the order Bunyavirales (Negarnaviricota: Polyploviricotina: Ellioviricetes) as of March 2020
References
- Abudurexiti A, Adkins S, Alioto D, Alkhovsky SV, Avšič-Županc T, Ballinger MJ, Bente DA, Beer M, Bergeron É, Blair CD, Briese T, Buchmeier MJ, Burt FJ, Calisher CH, Cháng C, Charrel RN, Choi IR, Clegg JCS, de la Torre JC, de Lamballerie X, Dèng F, Di Serio F, Digiaro M, Drebot MA, Duàn X, Ebihara H, Elbeaino T, Ergünay K, Fulhorst CF, Garrison AR, Gāo GF, Gonzalez J-PJ, Groschup MH, Günther S, Haenni A-L, Hall RA, Hepojoki J, Hewson R, Hú Z, Hughes HR, Jonson MG, Junglen S, Klempa B, Klingström J, Kòu C, Laenen L, Lambert AJ, Langevin SA, Liu D, Lukashevich IS, Luò T, Lǚ C, Maes P, de Souza WM, Marklewitz M, Martelli GP, Matsuno K, Mielke-Ehret N, Minutolo M, Mirazimi A, Moming A, Mühlbach H-P, Naidu R, Navarro B, Nunes MRT, Palacios G, Papa A, Pauvolid-Corrêa A, Pawęska JT, Qiáo J, Radoshitzky SR, Resende RO, Romanowski V, Sall AA, Salvato MS, Sasaya T, Shěn S, Shí X, Shirako Y, Simmonds P, Sironi M, Song J-W, Spengler JR, Stenglein MD, Sū Z, Sūn S, Táng S, Turina M, Wáng B, Wáng C, Wáng H, Wáng J, Wèi T, Whitfield AE, Zerbini FM, Zhāng J, Zhāng L, Zhāng Y, Zhang Y-Z, Zhāng Y, Zhou X, Zhū L, Kuhn JH (2019) Taxonomy of the order Bunyavirales: update 2019. Arch Virol 164:1949–1965
CAS PubMed Central Google Scholar - Afonso CL, Amarasinghe GK, Bányai K, Bào Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand F-X, Briese T, Bukreyev A, Calisher CH, Chandran K, Chéng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astúa J, Formenty P, Fouchier RA, Fù Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li C-X, Lin X-D, Liú L, Longdon B, Marton S, Maisner A, Mühlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Randall RE, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Shi M, Smither SJ, Stenglein MD, Stone DM, Takada A, Terregino C, Tesh RB, Tian J-H, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Verbeek M, Volchkov VE, Wahl-Jensen V, Walsh JA, Walker PJ, Wang D, Wang L-F, Wetzel T, Whitfield AE, Xiè JT, Yuen K-Y, Zhang Y-Z, Kuhn JH (2016) Taxonomy of the order Mononegavirales: update 2016. Arch Virol 161:2351–2360
CAS PubMed Central Google Scholar - Aitken TH, Woodall JP, De Andrade AHP, Bensabath G, Shope RE (1975) Pacui virus, phlebotomine flies, and small mammals in Brazil: an epidemiological study. Am J Trop Med Hyg 24:358–368
CAS Google Scholar - Alkan C, Alwassouf S, Piorkowski G, Bichaud L, Tezcan S, Dincer E, Ergunay K, Ozbel Y, Alten B, de Lamballerie X, Charrel RN (2015) Isolation, genetic characterization, and seroprevalence of Adana virus, a novel phlebovirus belonging to the Salehabad virus complex, in Turkey. J Virol 89:4080–4091
CAS PubMed Central Google Scholar - Alkan C, Erisoz Kasap O, Alten B, de Lamballerie X, Charrel RN (2016) Sandfly-borne phlebovirus isolations from Turkey: new insight into the Sandfly fever Sicilian and Sandfly fever Naples Species. PLoS Negl Trop Dis 10:e0004519
PubMed Central Google Scholar - Alkan C, Moin Vaziri V, Ayhan N, Badakhshan M, Bichaud L, Rahbarian N, Javadian E-A, Alten B, de Lamballerie X, Charrel RN (2017) Isolation and sequencing of Dashli virus, a novel Sicilian-like virus in sandflies from Iran; genetic and phylogenetic evidence for the creation of one novel species within the Phlebovirus genus in the Phenuiviridae family. PLoS Negl Trop Dis 11:e0005978
PubMed Central Google Scholar - Amarasinghe GK, Bào Y, Basler CF, Bavari S, Beer M, Bejerman N, Blasdell KR, Bochnowski A, Briese T, Bukreyev A, Calisher CH, Chandran K, Collins PL, Dietzgen RG, Dolnik O, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Fang Q, Formenty P, Fouchier RAM, Ghedin E, Harding RM, Hewson R, Higgins CM, Hong J, Horie M, James AP, Jiāng D, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lee B, Leroy EM, Li M, Maisner A, Mühlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Pearson MN, Randall RE, Revill PA, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Smither SJ, Song Q, Stone DM, Takada A, Terregino C, Tesh RB, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Volchkov VE, Wahl-Jensen V, Walker PJ, Wang B, Wang D, Wang F, Wang L-F, Werren JH, Whitfield AE, Yan Z, Ye G, Kuhn JH (2017) Taxonomy of the order Mononegavirales: update 2017. Arch Virol 162:2493–2504
CAS PubMed Central Google Scholar - Amarasinghe GK, Ceballos NGA, Banyard AC, Basler CF, Bavari S, Bennett AJ, Blasdell KR, Briese T, Bukreyev A, Caì Y, Calisher CH, Lawson CC, Chandran K, Chapman CA, Chiu CY, Choi K-S, Collins PL, Dietzgen RG, Dolja VV, Dolnik O, Domier LL, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Echevarría JE, Fooks AR, Formenty PBH, Fouchier RAM, Freuling CM, Ghedin E, Goldberg TL, Hewson R, Horie M, Hyndman TH, Jiāng D, Kityo R, Kobinger GP, Kondō H, Koonin EV, Krupovic M, Kurath G, Lamb RA, Lee B, Leroy EM, Maes P, Maisner A, Marston DA, Mor SK, Müller T, Mühlberger E, Ramírez VMN, Netesov SV, Ng TFF, Nowotny N, Palacios G, Patterson JL, Pawęska JT, Payne SL, Prieto K, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Siddell S, Smither SJ, Song Q, Song T, Stenglein MD, Stone DM, Takada A, Tesh RB, Thomazelli LM, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Vázquez-Morón S, Verdugo C, Volchkov VE, Wahl V, Walker PJ, Wang D, Wang L-F, Wellehan JFX, Wiley MR, Whitfield AE, Wolf YI, Yè G, Zhāng Y-Z, Kuhn JH (2018) Taxonomy of the order Mononegavirales: update 2018. Arch Virol 163:2283–2294
CAS PubMed Central Google Scholar - Amarasinghe GK, Ayllón MA, Bào Y, Basler CF, Bavari S, Blasdell KR, Briese T, Brown PA, Bukreyev A, Balkema-Buschmann A, Buchholz UJ, Chabi-Jesus C, Chandran K, Chiapponi C, Crozier I, de Swart RL, Dietzgen RG, Dolnik O, Drexler JF, Dürrwald R, Dundon WG, Duprex WP, Dye JM, Easton AJ, Fooks AR, Formenty PBH, Fouchier RAM, Freitas-Astúa J, Griffiths A, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondō H, Kurath G, Kuzmin IV, Lamb RA, Lavazza A, Lee B, Lelli D, Leroy EM, Lǐ J, Maes P, Marzano S-YL, Moreno A, Mühlberger E, Netesov SV, Nowotny N, Nylund A, Økland AL, Palacios G, Pályi B, Pawęska JT, Payne SL, Prosperi A, Ramos-González PL, Rima BK, Rota P, Rubbenstroth D, Shī M, Simmonds P, Smither SJ, Sozzi E, Spann K, Stenglein MD, Stone DM, Takada A, Tesh RB, Tomonaga K, Tordo N, Towner JS, van den Hoogen B, Vasilakis N, Wahl V, Walker PJ, Wang L-F, Whitfield AE, Williams JV, Zerbini FM, Zhāng T, Zhang Y-Z, Kuhn JH (2019) Taxonomy of the order Mononegavirales: update 2019. Arch Virol 164:1967–1980
CAS PubMed Central Google Scholar - Amaro F, Zé-Zé L, Alves MJ, Börstler J, Clos J, Lorenzen S, Becker SC, Schmidt-Chanasit J, Cadar D (2015) Co-circulation of a novel phlebovirus and Massilia virus in sandflies, Portugal. Virol J 12:174
PubMed Central Google Scholar - Barr J, Smith C, Smith I, de Jong C, Todd S, Melville D, Broos A, Crameri S, Haining J, Marsh G, Crameri G, Field H, Wang LF (2015) Isolation of multiple novel paramyxoviruses from pteropid bat urine. J Gen Virol 96:24–29
CAS Google Scholar - Bejerman N, de Breuil S, Debat H, Miretti M, Badaracco A, Nome C (2017) Molecular characterization of yerba mate chlorosis-associated virus, a putative cytorhabdovirus infecting yerba mate (Ilex paraguariensis). Arch Virol 162:2481–2484
CAS Google Scholar - Berge TO (1975) International Catalogue of Arboviruses Including Certain Other Viruses of Vertebrates. US Public Health Service publication no. (CDC) 75-8301, 2nd edn. Department of Health, Education and Welfare, Washington, DC
- Bhatt PN, Kulkarni KG, Boshell MJ, Rajagopalan PK, Patil AP, Goverdhan MK, Pavri KM (1966) Kaisodi virus, a new agent isolated from Haemaphysalis spinigera in Mysore State, South India. I. Isolation of strains. Am J Trop Med Hyg 15:958–960
CAS Google Scholar - Bichaud L, Dachraoui K, Alwassouf S, Alkan C, Mensi M, Piorkowski G, Sakhria S, Seston M, Fares W, De Lamballerie X, Zhioua E, Charrel RN (2016) Isolation, full genomic characterization and neutralization-based human seroprevalence of Medjerda Valley virus, a novel sandfly-borne phlebovirus belonging to the Salehabad virus complex in northern Tunisia. J Gen Virol 97:602–610
CAS Google Scholar - Bishop DHL, Pringle CR (1995) Order Mononegavirales. In: Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD (eds) Virus taxonomy—sixth report of the international committee on taxonomy of viruses/archives of virology supplement 10. Springer, Vienna, pp 265–267
Google Scholar - Bouquet J, Melgar M, Swei A, Delwart E, Lane RS, Chiu CY (2017) Metagenomic-based surveillance of Pacific Coast tick Dermacentor occidentalis identifies two novel bunyaviruses and an emerging human ricksettsial pathogen. Sci Rep 7:12234
PubMed Central Google Scholar - Buzkan N, Chiumenti M, Massart S, Sarpkaya K, Karadağ S, Minafra A (2019) A new emaravirus discovered in Pistacia from Turkey. Virus Res 263:159–163
CAS Google Scholar - Causey OR, Shope RE (1965) Icoaraci, a new virus related to Naples phlebotomus fever virus. Proc Soc Exp Biol Med 118:420–421
CAS Google Scholar - Charrel RN, Moureau G, Temmam S, Izri A, Marty P, Parola P, da Rosa AT, Tesh RB, de Lamballerie X (2009) Massilia virus, a novel Phlebovirus (Bunyaviridae) isolated from sandflies in the Mediterranean. Vector Borne Zoonotic Dis 9:519–530
PubMed Central Google Scholar - Chen CC, Chiu RJ (1996) A tospovirus infecting peanut in Taiwan. Acta Hortic 431:57–67
Google Scholar - Cheng Y-H, Zheng Y-X, Tai C-H, Yen J-H, Chen Y-K, Jan F-J (2014) Identification, characterisation and detection of a new tospovirus on sweet pepper. Ann Appl Biol 164:107–115
CAS Google Scholar - Coffey LL, Page BL, Greninger AL, Herring BL, Russell RC, Doggett SL, Haniotis J, Wang C, Deng X, Delwart EL (2014) Enhanced arbovirus surveillance with deep sequencing: identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes. Virology 448:146–158
CAS Google Scholar - de Carvalho MS, de Lara Pinto AZ, Pinheiro A, Rodrigues JSV, Melo FL, da Silva LA, Ribeiro BM, Dezengrini-Slhessarenko R (2018) Viola phlebovirus is a novel Phlebotomus fever serogroup member identified in Lutzomyia (Lutzomyia) longipalpis from Brazilian Pantanal. Parasit Vectors 11:405
PubMed Central Google Scholar - De Haas RA, Jonkers AH, Heinemann DW (1966) Kwatta virus, a new agent isolated from Culex mosquitoes in Surinam. Am J Trop Med Hyg 15:954–957
Google Scholar - Debat HJ, Bejerman N (2019) Novel bird’s-foot trefoil RNA viruses provide insights into a clade of legume-associated enamoviruses and rhabdoviruses. Arch Virol 164:1419–1426
CAS Google Scholar - Dilcher M, Alves MJ, Finkeisen D, Hufert F, Weidmann M (2012) Genetic characterization of Bhanja virus and Palma virus, two tick-borne phleboviruses. Virus Genes 45:311–315
CAS Google Scholar - Dilcher M, Faye O, Faye O, Weber F, Koch A, Sadegh C, Weidmann M, Sall AA (2015) Zahedan rhabdovirus, a novel virus detected in ticks from Iran. Virol J 12:183
PubMed Central Google Scholar - Doherty RL, Carley JG, Standfast HA, Dyce AL, Kay BH, Snowdon WA (1973) Isolation of arboviruses from mosquitoes, biting midges, sandflies and vertebrates collected in Queensland, 1969 and 1970. Trans R Soc Trop Med Hyg 67:536–543
CAS Google Scholar - Dong J-H, Cheng X-F, Yin YY, Fang Q, Ding M, Li T-T, Zhang L-Z, Su X-X, McBeath J-H, Zhang Z-K (2008) Characterization of tomato zonate spot virus, a new tospovirus in China. Arch Virol 153:855–864
CAS Google Scholar - Dong JH, Yin YY, Fang Q, McBeath JH, Zhang ZK (2013) A new tospovirus causing chlorotic ringspot on Hippeastrum sp. in China. Virus Genes 46:567–570
CAS Google Scholar - Easton AJ, Pringle CR (2011) Order Mononegavirales. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy—ninth report of the international committee on taxonomy of viruses. Elsevier/Academic Press, London, pp 653–657
Google Scholar - Fernandes J, Guterres A, de Oliveira RC, Chamberlain J, Lewandowski K, Teixeira BR, Coelho TA, Crisóstomo CF, Bonvicino CR, D’Andrea PS, Hewson R, de Lemos ERS (2018) Xapuri virus, a novel mammarenavirus: natural reassortment and increased diversity between New World viruses. Emerg Microbes Infect 7:120
PubMed Central Google Scholar - Fernandes J, Guterres A, de Oliveira RC, Jardim R, Dávila AMR, Hewson R, de Lemos ERS (2019) Apore virus, a novel mammarenavirus (Bunyavirales: Arenaviridae) related to highly pathogenic virus from South America. Mem Inst Oswaldo Cruz 114:e180586
PubMed Central Google Scholar - Forth LF, Konrath A, Klose K, Schlottau K, Hoffmann K, Ulrich RG, Höper D, Pohlmann A, Beer M (2018) A novel squirrel respirovirus with putative zoonotic potential. Viruses 10:373
Google Scholar - Gaafar YZA, Richert-Pöggeler KR, Maaß C, Vetten H-J, Ziebell H (2019) Characterisation of a novel nucleorhabdovirus infecting alfalfa (Medicago sativa). Virol J 16:55
PubMed Central Google Scholar - Goldstein T, Anthony SJ, Gbakima A, Bird BH, Bangura J, Tremeau-Bravard A, Belaganahalli MN, Wells HL, Dhanota JK, Liang E, Grodus M, Jangra RK, DeJesus VA, Lasso G, Smith BR, Jambai A, Kamara BO, Kamara S, Bangura W, Monagin C, Shapira S, Johnson CK, Saylors K, Rubin EM, Chandran K, Lipkin WI, Mazet JAK (2018) The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol 3:1084–1089
CAS PubMed Central Google Scholar - Gubala A, Walsh S, McAllister J, Weir R, Davis S, Melville L, Mitchell I, Bulach D, Gauci P, Skvortsov A, Boyle D (2017) Identification of very small open reading frames in the genomes of Holmes Jungle virus, Ord River virus, and Wongabel virus of the genus Hapavirus, family Rhabdoviridae. Evol Bioinform Online 13:1176934317713484
PubMed Central Google Scholar - Hang J, Klein TA, Kim H-C, Yang Y, Jima DD, Richardson JH, Jarman RG (2016) Genome sequences of five arboviruses in field-captured mosquitoes in a unique rural environment of South Korea. Genome Announc 4:e01644-15
PubMed Central Google Scholar - Hannoun C, Corniou B, Rageau J (1970) Isolation in southern France and characterization of new tick-borne viruses related to Uukuniemi: Grand Arbaud and Ponteves. Acta Virol 14:167–170
CAS Google Scholar - Hassan M, Di Bello PL, Keller KE, Martin RR, Sabanadzovic S, Tzanetakis IE (2017) A new, widespread emaravirus discovered in blackberry. Virus Res 235:1–5
CAS Google Scholar - Hassani-Mehraban A, Botermans M, Verhoeven JTJ, Meekes E, Saaijer J, Peters D, Goldbach R, Kormelink R (2010) A distinct tospovirus causing necrotic streak on Alstroemeria sp. in Colombia. Arch Virol 155:423–428
CAS PubMed Central Google Scholar - Hassani-Mehraban A, Dullemans AM, Verhoeven JTJ, Roenhorst JW, Peters D, van der Vlugt RAA, Kormelink R (2019) Alstroemeria yellow spot virus (AYSV): a new orthotospovirus species within a growing Eurasian clade. Arch Virol 164:117–126
CAS Google Scholar - Hepojoki J, Hepojoki S, Smura T, Szirovicza L, Dervas E, Prahauser B, Nufer L, Schraner EM, Vapalahti O, Kipar A, Hetzel U (2018) Characterization of Haartman Institute snake virus-1 (HISV-1) and HISV-like viruses - the representatives of genus Hartmanivirus, family Arenaviridae. PLoS Pathog 14:e1007415
PubMed Central Google Scholar - Hu S-C, Hsu C-L, Lee M-S, Tu Y-C, Chang J-C, Wu C-H, Lee S-H, Ting L-J, Tsai K-R, Cheng M-C, Tu W-J, Hsu W-C (2018) Lyssavirus in Japanese pipistrelle, Taiwan. Emerg Infect Dis 24:782–785
CAS PubMed Central Google Scholar - Hughes HR, Russell BJ, Lambert AJ (2020) Genetic characterization of Frijoles and Chilibre species complex viruses (genus Phlebovirus; family Phenuiviridae) and three unclassified New World phleboviruses. Am J Trop Med Hyg 102:359–365
CAS Google Scholar - Ito T, Suzaki K, Nakano M (2013) Genetic characterization of novel putative rhabdovirus and dsRNA virus from Japanese persimmon. J Gen Virol 94:1917–1921
CAS Google Scholar - Jeong J, Kim Y, An I, Wang S-J, Kim Y, Lee H-J, Choi K-S, Im S-P, Min W, Oem J-K, Jheong W (2018) Complete genome sequence of a novel avian paramyxovirus isolated from wild birds in South Korea. Arch Virol 163:223–227
CAS Google Scholar - Johnson RI, Tachedjian M, Rowe B, Clayton BA, Layton R, Bergfeld J, Wang L-F, Marsh GA (2018) Alston virus, a novel paramyxovirus isolated from bats causes upper respiratory tract infection in experimentally challenged ferrets. Viruses 10:675
CAS Google Scholar - Jones S, McGavin W, MacFarlane S (2019) The complete sequences of two divergent variants of the rhabdovirus raspberry vein chlorosis virus and the design of improved primers for virus detection. Virus Res 265:162–165
CAS Google Scholar - Karabatsos N (1985) International catalogue of arboviruses including certain other viruses of vertebrates. American Society for Tropical Medicine and Hygiene, San Antonio
Google Scholar - Kerschner JH, Calisher CH, Vorndam AV, Francy DB (1986) Identification and characterization of Bahia Grande, Reed Ranch and Muir Springs viruses, related members of the family Rhabdoviridae with widespread distribution in the United States. J Gen Virol 67:1081–1089
CAS Google Scholar - Kohl C, Tachedjian M, Todd S, Monaghan P, Boyd V, Marsh GA, Crameri G, Field H, Kurth A, Smith I, Wang L-F (2018) Hervey virus: a study on co-circulation with henipaviruses in pteropid bats within their distribution range from Australia to Africa. PLoS One 13:e0191933
PubMed Central Google Scholar - Kokernot RH, Calisher CH, Stannard LJ, Hayes J (1969) Arbovirus studies in the Ohio-Mississippi Basin, 1964–1967. VII. Lone star virus, a hitherto unknown agent isolated from the tick Amblyomma americanum (Linn.). Am J Trop Med Hyg 18:789–795
CAS Google Scholar - Koonin EV, Dolja VV, Krupovic M, Arvind V, Wolf YI, Yutin N, Zerbini FM, Kuhn JH (2020) Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev 84:e00061-19
Google Scholar - Kuhn JH, Wolf YI, Krupovic M, Zhang Y-Z, Maes P, Dolja VV, Koonin EV (2019) Classify viruses—the gain is worth the pain. Nature 566:318–320
Google Scholar - Lecoq H, Wipf-Scheibel C, Verdin E, Desbiez C (2019) Characterization of the first tenuivirus naturally infecting dicotyledonous plants. Arch Virol 164:297–301
CAS Google Scholar - Ledermann JP, Zeidner N, Borland EM, Mutebi J-P, Lanciotti RS, Miller BR, Lutwama JJ, Tendo JM, Andama V, Powers AM (2014) Sunguru virus: a novel virus in the family Rhabdoviridae isolated from a chicken in north-western Uganda. J Gen Virol 95:1436–1443
CAS PubMed Central Google Scholar - Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, Qin X-C, Xu J, Holmes EC, Zhang Y-Z (2015) Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4:e05378
Google Scholar - Lin Y-H, Fujita M, Chiba S, Hyodo K, Andika IB, Suzuki N, Kondo H (2019) Two novel fungal negative-strand RNA viruses related to mymonaviruses and phenuiviruses in the shiitake mushroom (Lentinula edodes). Virology 533:125–136
CAS Google Scholar - Liu Y, Du Z, Wang H, Zhang S, Cao M, Wang X (2018) Identification and characterization of wheat yellow striate virus, a novel leafhopper-transmitted nucleorhabdovirus infecting wheat. Front Microbiol 9:468
PubMed Central Google Scholar - Lvov DK, Timopheeva AA, Gromashevski VL, Gostinshchikova GV, Veselovskaya OV, Chervonski VI, Fomina KB, Gromov AI, Pogrebenko AG, Zhezmer VY (1973) “Zaliv Terpeniya” virus, a new Uukuniemi group arbovirus isolated from Ixodes (Ceratixodes) putus Pick.-Camb. 1878 on Tyuleniy Island (Sakhalin region) and Commodore Islands (Kamchatsk region). Arch Gesamte Virusforsch 41:165–169
CAS Google Scholar - Lvov SD, Gromashevsky VL, Andreev VP, Skvortsova TM, Kondrashina NG, Morozova TN, Avershin AD, Aristova VA, Dmitriev GA, Kandaurov YK, Kuznetsov AA, Galkina IV, Yamnikova SS, Shchipanova MV (1990) Natural foci of arboviruses in far northern latitudes of Eurasia. In: Calisher CH (ed) Hemorrhagic fever with renal syndrome, tick- and mosquito-borne viruses. Springer, Vienna, pp 267–275
Google Scholar - Maes P, Alkhovsky SV, Bào Y, Beer M, Birkhead M, Briese T, Buchmeier MJ, Calisher CH, Charrel RN, Choi IR, Clegg CS, Torre JC, Delwart E, DeRisi JL, Bello PLD, Serio FD, Digiaro M, Dolja VV, Drosten C, Druciarek TZ, Du J, Ebihara H, Elbeaino T, Gergerich RC, Gillis AN, Gonzalez J-PJ, Haenni A-L, Hepojoki J, Hetzel U, Hồ T, Hóng N, Jain RK, Vuren PJv, Jin Q, Jonson MG, Junglen S, Keller KE, Kemp A, Kipar A, Kondov NO, Koonin EV, Kormelink R, Korzyukov Y, Krupovic M, Lambert AJ, Laney AG, LeBreton M, Lukashevic IS, Marklewitz M, Markotter W, Martelli GP, Martin RR, Mielke-Ehret N, Mühlbach H-P, Navarro B, Ng TFF, Nunes MRT, Palacios G, Pawęska JT, Peters CJ, Plyusnin A, Radoshitzky SR, Romanowski V, Salmenperä P, Salvato MS, Sanfaçon H, Sasaya T, Schmaljohn C, Schneider BS, Shirako Y, Siddell S, Sironen TA, Stenglein MD, Storm N, Sudini H, Tesh RB, Tzanetakis IE, Uppala M, Vapalahti O, Vasilakis N, Walker PJ, Wáng G, Wáng L, Wáng Y, Wèi T, Wiley MR, Wolf YI, Wolfe ND, Wú Z, Xú W, Yang L, Yāng Z, Yeh S-D, Zhāng Y-Z, Zhèng Y, Zhou X, Zhū C, Zirkel F, Kuhn JH (2018) Taxonomy of the family Arenaviridae and the order Bunyavirales: update 2018. Arch Virol 163:2295–310
- Maes P, Adkins S, Alkhovsky SV, Avšič-Županc T, Ballinger MJ, Bente DA, Beer M, Bergeron É, Blair CD, Briese T, Buchmeier MJ, Burt FJ, Calisher CH, Charrel RN, Choi IR, Clegg JCS, de la Torre JC, de Lamballerie X, DeRisi JL, Digiaro M, Drebot M, Ebihara H, Elbeaino T, Ergünay K, Fulhorst CF, Garrison AR, Gāo GF, Gonzalez J-PJ, Groschup MH, Günther S, Haenni A-L, Hall RA, Hewson R, Hughes HR, Jain RK, Jonson MG, Junglen S, Klempa B, Klingström J, Kormelink R, Lambert AJ, Langevin SA, Lukashevich IS, Marklewitz M, Martelli GP, Mielke-Ehret N, Mirazimi A, Mühlbach H-P, Naidu R, Nunes MRT, Palacios G, Papa A, Pawęska JT, Peters CJ, Plyusnin A, Radoshitzky SR, Resende RO, Romanowski V, Sall AA, Salvato MS, Sasaya T, Schmaljohn C, Shí X, Shirako Y, Simmonds P, Sironi M, Song J-W, Spengler JR, Stenglein MD, Tesh RB, Turina M, Wèi T, Whitfield AE, Yeh S-D, Zerbini FM, Zhang Y-Z, Zhou X, Kuhn JH (2019) Taxonomy of the order Bunyavirales: second update 2018. Arch Virol 164:927–41
- Maes P, Amarasinghe GK, Ayllón MA, Basler CF, Bavari S, Blasdell KR, Briese T, Brown PA, Bukreyev A, Balkema-Buschmann A, Buchholz UJ, Chandran K, Crozier I, de Swart RL, Dietzgen RG, Dolnik O, Domier LL, Drexler JF, Dürrwald R, Dundon WG, Duprex WP, Dye JM, Easton AJ, Fooks AR, Formenty PBH, Fouchier RAM, Freitas-Astúa J, Ghedin E, Griffiths A, Hewson R, Horie M, Hurwitz JL, Hyndman TH, Jiāng D, Kobinger GP, Kondō H, Kurath G, Kuzmin IV, Lamb RA, Lee B, Leroy EM, Lǐ J, Marzano S-YL, Mühlberger E, Netesov SV, Nowotny N, Palacios G, Pályi B, Pawęska JT, Payne SL, Rima BK, Rota P, Rubbenstroth D, Simmonds P, Smither SJ, Song Q, Song T, Spann K, Stenglein MD, Stone DM, Takada A, Tesh RB, Tomonaga K, Tordo N, Towner JS, van den Hoogen B, Vasilakis N, Wahl V, Walker PJ, Wang D, Wang L-F, Whitfield AE, Williams JV, Yè G, Zerbini FM, Zhang Y-Z, Kuhn JH (2019) Taxonomy of the order Mononegavirales: second update 2018. Arch Virol 164:1233–1244
CAS PubMed Central Google Scholar - Main AJ, Carey AB (1980) Connecticut virus: a new Sawgrass group virus from Ixodes dentatus (Acari: Ixodidae). J Med Entomol 17:473–476
Google Scholar - Marklewitz M, Dutari LC, Paraskevopoulou S, Page RA, Loaiza JR, Junglen S (2019) Diverse novel phleboviruses in sandflies from the Panama Canal area, Central Panama. J Gen Virol 100:938–949
CAS Google Scholar - Matsuno K, Weisend C, Kajihara M, Matysiak C, Williamson BN, Simuunza M, Mweene AS, Takada A, Tesh RB, Ebihara H (2015) Comprehensive molecular detection of tick-borne phleboviruses leads to the retrospective identification of taxonomically unassigned bunyaviruses and the discovery of a novel member of the genus Phlebovirus. J Virol 89:594–604
CAS Google Scholar - Maurino F, Dumón AD, Llauger G, Alemandri V, de Haro LA, Mattio MF, Del Vas M, Laguna IG, MdlP Giménez Pecci (2018) Complete genome sequence of maize yellow striate virus, a new cytorhabdovirus infecting maize and wheat crops in Argentina. Arch Virol 163:291–295
CAS Google Scholar - McAllister J, Gauci PJ, Mitchell IR, Boyle DB, Bulach DM, Weir RP, Melville LF, Davis SS, Gubala AJ (2014) Genomic characterisation of Almpiwar virus, Harrison Dam virus and Walkabout Creek virus; three novel rhabdoviruses from northern Australia. Virol Rep 3–4:1–17
Google Scholar - McLean DM, Larke RPB (1963) Powassan and Silverwater viruses: ecology of two Ontario arboviruses. Can Med Assoc J 88:182–185
CAS PubMed Central Google Scholar - Medina-Salguero AX, Cornejo-Franco JF, Grinstead S, Mollov D, Mowery JD, Flores F, Quito-Avila DF (2019) Sequencing, genome analysis and prevalence of a cytorhabdovirus discovered in Carica papaya. PLoS One 14:e0215798
CAS PubMed Central Google Scholar - Meng J, Liu P, Zhu L, Zou C, Li J, Chen B (2015) Complete genome sequence of mulberry vein banding associated virus, a new tospovirus infecting mulberry. PLoS One 10:e0136196
PubMed Central Google Scholar - Meng JR, Liu PP, Zou CW, Wang ZQ, Liao YM, Cai JH, Qin BX, Chen BS (2013) First report of a Tospovirus in mulberry. Plant Dis 97:1001
CAS Google Scholar - Menzel W, Richert-Pöggeler KR, Winter S, Knierim D (2018) Characterization of a nucleorhabdovirus from Physostegia. Acta Hortic 1193:29–38
Google Scholar - Muller MJ, Standfast HA (1986) Vectors of ephemeral fever group viruses. In: St George TD, Kay BH, Blok J (eds) Arbovirus research in Australia—proceedings of the fourth symposium. CSIRO/QMIR, Brisbane, pp 295–300
- Navarro B, Zicca S, Minutolo M, Saponari M, Alioto D, Di Serio F (2018) A negative-stranded RNA virus infecting citrus trees: the second member of a new genus within the order Bunyavirales. Front Microbiol 9:2340
PubMed Central Google Scholar - Noh JY, Jeong DG, Yoon S-W, Kim JH, Choi YG, Kang S-Y, Kim HK (2018) Isolation and characterization of novel bat paramyxovirus B16-40 potentially belonging to the proposed genus Shaanvirus. Sci Rep 8:12533
PubMed Central Google Scholar - Nunes-Neto JP, Souza WM, Acrani GO, Romeiro MF, Fumagalli M, Vieira LC, Medeiros DBdA, Lima JA, de Lima CPS, Cardoso JF, Figueiredo LTM, da Silva SPD, Tesh R, Nunes MRT, Vasconcelos PFdC (2017) Characterization of the Bujaru, frijoles and Tapara antigenic complexes into the sandfly fever group and two unclassified phleboviruses from Brazil. J Gen Virol 98:585–594
CAS Google Scholar - Økland AL, Nylund A, Øvergård A-C, Skoge RH, Kongshaug H (2019) Genomic characterization, phylogenetic position and in situ localization of a novel putative mononegavirus in Lepeophtheirus salmonis. Arch Virol 164:675–689
Google Scholar - Palacios G, da Rosa AT, Savji N, Sze W, Wick I, Guzman H, Hutchison S, Tesh R, Lipkin WI (2011) Aguacate virus, a new antigenic complex of the genus Phlebovirus (family Bunyaviridae). J Gen Virol 92:1445–1453
CAS PubMed Central Google Scholar - Palacios G, Tesh R, Travassos da Rosa A, Savji N, Sze W, Jain K, Serge R, Guzman H, Guevara C, Nunes MR, Nunes-Neto JP, Kochel T, Hutchison S, Vasconcelos PFC, Lipkin WI (2011) Characterization of the Candiru antigenic complex (Bunyaviridae: Phlebovirus), a highly diverse and reassorting group of viruses affecting humans in tropical America. J Virol 85:3811–3820
CAS PubMed Central Google Scholar - Palacios G, Savji N, Travassos da Rosa A, Desai A, Sanchez-Seco MP, Guzman H, Lipkin WI, Tesh R (2013) Characterization of the Salehabad virus species complex of the genus Phlebovirus (Bunyaviridae). J Gen Virol 94:837–842
CAS PubMed Central Google Scholar - Palacios G, Savji N, Travassos da Rosa A, Guzman H, Yu X, Desai A, Rosen GE, Hutchison S, Lipkin WI, Tesh R (2013) Characterization of the Uukuniemi virus group (Phlebovirus: Bunyaviridae): evidence for seven distinct species. J Virol 87:3187–3195
CAS PubMed Central Google Scholar - Palacios G, Tesh RB, Savji N, Travassos da Rosa APA, Guzman H, Bussetti AV, Desai A, Ladner J, Sanchez-Seco M, Lipkin WI (2014) Characterization of the Sandfly fever Naples species complex and description of a new Karimabad species complex (genus Phlebovirus, family Bunyaviridae). J Gen Virol 95:292–300
CAS PubMed Central Google Scholar - Palacios G, Wiley MR, Travassos da Rosa APA, Guzman H, Quiroz E, Savji N, Carrera J-P, Bussetti AV, Ladner JT, Lipkin WI, Tesh RB (2015) Characterization of the Punta Toro species complex (genus Phlebovirus, family Bunyaviridae). J Gen Virol 96:2079–2085
CAS PubMed Central Google Scholar - Pavri KM, Casals J (1966) Kaisodi virus, a new agent isolated from Haemaphysalis spinigera in Mysore state, South India. Am J Trop Med Hyg 15:961–963
CAS Google Scholar - Pecman A, Kutnjak D, Gutiérrez-Aguirre I, Adams I, Fox A, Boonham N, Ravnikar M (2017) Next generation sequencing for detection and discovery of plant viruses and viroids: comparison of two approaches. Front Microbiol 8:1998
PubMed Central Google Scholar - Peralta PH, Shelokov A, Brody JA (1965) Chagres virus: a new human isolate from Panama. Am J Trop Med Hyg 14:146–151
CAS Google Scholar - Pettersson JH-O, Shi M, Bohlin J, Eldholm V, Brynildsrud OB, Paulsen KM, Andreassen Å, Holmes EC (2017) Characterizing the virome of Ixodes ricinus ticks from northern Europe. Sci Rep 7:10870
PubMed Central Google Scholar - Pringle CR, Alexander DJ, Billeter MA, Collins PL, Kingsbury DW, Lipkind MA, Nagai Y, Orvell C, Rima B, Rott R, ter Meulen V (1991) The order Mononegavirales. Arch Virol 117:137–140
Google Scholar - Pringle CR (1997) The order _Mononegavirales_—current status. Arch Virol 142:2321–2326
CAS Google Scholar - Pringle CR (2000) Order Mononegavirales. In: van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB (eds) Virus taxonomy—seventh report of the international committee on taxonomy of viruses. Academic Press, San Diego, pp 525–530
Google Scholar - Pringle CR (2005) Order Mononegavirales. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy—eighth report of the international committee on taxonomy of viruses. Elsevier/Academic Press, San Diego, pp 609–614
Google Scholar - Quan P-L, Williams DT, Johansen CA, Jain K, Petrosov A, Diviney SM, Tashmukhamedova A, Hutchison SK, Tesh RB, Mackenzie JS, Briese T, Lipkin WI (2011) Genetic characterization of K13965, a strain of Oak Vale virus from Western Australia. Virus Res 160:206–213
CAS PubMed Central Google Scholar - Read DA, Featherston J, Rees DJG, Thompson GD, Roberts R, Flett BC, Mashingaidze K, Pietersen G, Kiula B, Kullaya A, Mbega ER (2019) Molecular characterization of Morogoro maize-associated virus, a nucleorhabdovirus detected in maize (Zea mays) in Tanzania. Arch Virol 164:1711–1715
CAS Google Scholar - Reuter G, Boros A, Pál J, Kapusinszky B, Delwart E, Pankovics P (2016) Detection and genome analysis of a novel (dima)rhabdovirus (Riverside virus) from Ochlerotatus sp. mosquitoes in Central Europe. Infect Genet Evol 39:336–341
CAS Google Scholar - Ritter DG, Calisher CH, Muth DJ, Shope RE, Murphy FA, Whitfield SG (1978) New Minto virus: a new rhabdovirus from ticks in Alaska. Can J Microbiol 24:422–426
CAS Google Scholar - Rott ME, Kesanakurti P, Berwarth C, Rast H, Boyes I, Phelan J, Jelkmann W (2018) Discovery of negative-sense RNA viruses in trees infected with apple rubbery wood disease by next-generation sequencing. Plant Dis 102:1254–1263
CAS Google Scholar - Sabin AB (1951) Experimental studies on Phlebotomus (pappataci, sandfly) fever during World War II. Arch Gesamte Virusforsch 4:367–410
CAS Google Scholar - Sather GE, Lewis AL, Jennings W, Bond JO, Hammon WM (1970) Sawgrass virus: a newly described arbovirus in Florida. Am J Trop Med Hyg 19:319–326
CAS Google Scholar - Scarpassa VM, Debat HJ, Alencar RB, Saraiva JF, Calvo E, Arcà B, Ribeiro JMC (2019) An insight into the sialotranscriptome and virome of Amazonian anophelines. BMC Genom 20:166
Google Scholar - Shah KV, Work TH (1969) Bhanja virus: a new arbovirus from ticks Haemaphysalis intermedia Warburton and Nuttall, 1909, in Orissa, India. Indian J Med Res 57:793–798
CAS Google Scholar - Shahhosseini N, Lühken R, Jöst H, Jansen S, Börstler J, Rieger T, Krüger A, Yadouleton A, de Mendonça Campos R, Cirne-Santos CC, Ferreira DF, Garms R, Becker N, Tannich E, Cadar D, Schmidt-Chanasit J (2017) Detection and characterization of a novel rhabdovirus in Aedes cantans mosquitoes and evidence for a mosquito-associated new genus in the family Rhabdoviridae. Infect Genet Evol 55:260–268
Google Scholar - Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, Buchmann J, Wang W, Xu J, Holmes EC, Zhang YZ (2016) Redefining the invertebrate RNA virosphere. Nature 540:539–543
CAS Google Scholar - Shi M, Neville P, Nicholson J, Eden J-S, Imrie A, Holmes EC (2017) High-resolution metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in western Australia. J Virol 91:e00680-17
PubMed Central Google Scholar - Shi M, Lin X-D, Chen X, Tian J-H, Chen L-J, Li K, Wang W, Eden J-S, Shen J-J, Liu L, Holmes EC, Zhang Y-Z (2018) The evolutionary history of vertebrate RNA viruses. Nature 556:197–202
CAS Google Scholar - Siddell SG, Walker PJ, Lefkowitz EJ, Mushegian AR, Adams MJ, Dutilh BE, Gorbalenya AE, Bz Harrach, Harrison RL, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert M, Rubino L, Sabanadzovic S, Sanfaçon H, Simmonds P, Varsani A, Zerbini FM, Davison AJ (2019) Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018). Arch Virol 164:943–946
CAS Google Scholar - Spence L, Anderson CR, Aitken THG, Downs WG (1966) Aruac virus, a new agent isolated from Trinidadian mosquitoes. Am J Trop Med Hyg 15:231–234
CAS Google Scholar - St George TD, Doherty RL, Carley JG, Filippich C, Brescia A, Casals J, Kemp DH, Brothers N (1985) The isolation of arboviruses including a new flavivirus and a new bunyavirus from Ixodes (Ceratixodes) uriae (Ixodoidea: Ixodidae) collected at Macquarie Island, Australia, 1975–1979. Am J Trop Med Hyg 34:406–412
CAS Google Scholar - Straková P, Dufkova L, Širmarová J, Salát J, Bartonička T, Klempa B, Pfaff F, Höper D, Hoffmann B, Ulrich RG, Růžek D (2017) Novel hantavirus identified in European bat species Nyctalus noctula. Infect Genet Evol 48:127–130
Google Scholar - Sun Q, Zhao Q, An X, Guo X, Zuo S, Zhang X, Pei G, Liu W, Cheng S, Wang Y, Shu P, Mi Z, Huang Y, Zhang Z, Tong Y, Zhou H, Zhang J (2017) Complete genome sequence of Menghai rhabdovirus, a novel mosquito-borne rhabdovirus from China. Arch Virol 162:1103–1106
CAS Google Scholar - Swei A, Russell BJ, Naccache SN, Kabre B, Veeraraghavan N, Pilgard MA, Johnson BJB, Chiu CY (2013) The genome sequence of lone star virus, a highly divergent bunyavirus found in the Amblyomma americanum tick. PLoS One 8:e62083
CAS PubMed Central Google Scholar - Tchouassi DP, Marklewitz M, Chepkorir E, Zirkel F, Agha SB, Tigoi CC, Koskei E, Drosten C, Borgemeister C, Torto B, Junglen S, Sang R (2019) Sand fly-associated phlebovirus with evidence of neutralizing antibodies in humans, Kenya. Emerg Infect Dis 25:681–690
CAS PubMed Central Google Scholar - Tesh RB, Chaniotis BN, Peralta PH, Johnson KM (1974) Ecology of viruses isolated from Panamanian phlebotomine sandflies. Am J Trop Med Hyg 23:258–269
CAS Google Scholar - Tesh RB (1975) Multiplication of Phlebotomus fever group arboviruses in mosquitos after intrathoracic inoculation. J Med Entomol 12:1–4
CAS Google Scholar - Tesh RB, Boshell J, Young DG, Morales A, De Carrasquilla CF, Corredor A, Modi GB, Travassos Da Rosa APA, McLean RG, De Rodriguez C, Gaitan MO (1989) Characterization of 5 new phleboviruses recently isolated from sand flies in tropical America. Am J Trop Med Hyg 40:529–533
CAS Google Scholar - Tokarz R, Sameroff S, Leon MS, Jain K, Lipkin WI (2014) Genome characterization of Long Island tick rhabdovirus, a new virus identified in Amblyomma americanum ticks. Virol J 11:26
PubMed Central Google Scholar - Tokarz R, Williams SH, Sameroff S, Sanchez Leon M, Jain K, Lipkin WI (2014) Virome analysis of Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks reveals novel highly divergent vertebrate and invertebrate viruses. J Virol 88:11480–11492
PubMed Central Google Scholar - Trapp EE, Paes de Andrade AH, Shope RE (1965) Itaporanga, a newly reecognized arbovirus from Sao Paulo State, Brazil. Proc Soc Exp Biol Med 118:421–422
CAS Google Scholar - Travassos da Rosa APA, Tesh RB, Pinheiro FP, Travassos da Rosa JFS, Peterson NE (1983) Characterization of eight new phlebotomus fever serogroup arboviruses (Bunyaviridae: Phlebovirus) from the Amazon region of Brazil. Am J Trop Med Hyg 32:1164–1171
CAS Google Scholar - Vasilakis N, Tesh RB, Widen SG, Mirchandani D, Walker PJ (2019) Genomic characterisation of Cuiaba and Charleville viruses: arboviruses (family Rhabdoviridae, genus Sripuvirus) infecting reptiles and amphibians. Virus Genes 55:87–94
CAS Google Scholar - Velasco L, Arjona-Girona I, Cretazzo E, López-Herrera C (2019) Viromes in Xylariaceae fungi infecting avocado in Spain. Virology 532:11–21
CAS Google Scholar - Verani P, Ciufolini MG, Nicoletti L, Balducci M, Sabatinelli G, Coluzzi M, Paci P, Amaducci L (1982) Studi ecologici ed epidemiologici del virus Toscana, un arbovirus isolato da flebotomi. Ann Ist Super Sanita 18:397–9
- Walker PJ, Firth C, Widen SG, Blasdell KR, Guzman H, Wood TG, Paradkar PN, Holmes EC, Tesh RB, Vasilakis N (2015) Evolution of genome size and complexity in the Rhabdoviridae. PLoS Pathog 11:e1004664
PubMed Central Google Scholar - Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Dempsey DM, Dutilh BE, Harrach B, Harrison RL, Hendrickson RC, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert M, Rubino L, Sabanadzovic S, Simmonds P (2019) Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch Virol 164:2417–2429
CAS Google Scholar - Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens E, Dempsey DM, Dutilh BE, Harrach Bz, Harrison RL, Hendrickson RC, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert M, Rubino L, Sabanadzovic S, Simmonds P, Varsani A, Zerbini FM, Davison AJ (2020) Changes to virus taxonomy and the Statutes ratified by the International Committee on Taxonomy of Viruses (2020). Arch Virol
- Wang H, Liu Y, Liu W, Cao M, Wang X (2019) Sequence analysis and genomic organization of a novel chuvirus, Tàiyuán leafhopper virus. Arch Virol 164:617–620
CAS Google Scholar - Wang J, Selleck P, Yu M, Ha W, Rootes C, Gales R, Wise T, Crameri S, Chen H, Broz I, Hyatt A, Woods R, Meehan B, McCullough S, Wang L-F (2014) Novel phlebovirus with zoonotic potential isolated from ticks, Australia. Emerg Infect Dis 20:1040–1043
PubMed Central Google Scholar - Wang Y, Hua W, Wang J, Hannoufa A, Xu Z, Wang Z (2013) Deep sequencing of Lotus corniculatus L. reveals key enzymes and potential transcription factors related to the flavonoid biosynthesis pathway. Mol Genet Genom 288:131–139
CAS Google Scholar - Wanzeller ALM, Martins LC, Diniz Júnior JAP, de Almeida Medeiros DB, Cardoso JF, da Silva DEA, de Oliveira LF, de Vasconcelos JM, Nunes MRT, da S. G. Vianez Júnior JL, Vasconcelos PFC (2014) Xiburema virus, a hitherto undescribed virus within the family Rhabdoviridae isolated in the Brazilian Amazon Region. Genome Announc 2:e00454-14
PubMed Central Google Scholar - Willie K, Stewart LR (2017) Complete genome sequence of a new maize-associated cytorhabdovirus. Genome Announc 5:e00591-17
PubMed Central Google Scholar - Winter S, Koerbler M, Shahraeen N, Katul L, Lesemann D-E (2003) Characterization of a new tospovirus species infecting tomato in Iran. J Plant Dis Prot 110:74
Google Scholar - Winton JR, Batts WN, Powers RL, Purcell MK (2019) Complete genome sequences of the index isolates of two genotypes of Pacific salmon paramyxovirus. Microbiol Resour Announc 8:e01521-18
PubMed Central Google Scholar - Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV (2018) Origins and evolution of the global RNA virome. MBio 9:e02329-18
PubMed Central Google Scholar - Woyessa AB, Omballa V, Wang D, Lambert A, Waiboci L, Ayele W, Ahmed A, Abera NA, Cao S, Ochieng M, Montgomery JM, Jima D, Fields B (2014) An outbreak of acute febrile illness caused by sandfly fever Sicilian virus in the Afar Region of Ethiopia, 2011. Am J Trop Med Hyg 91:1250–1253
PubMed Central Google Scholar - Wu L-P, Yang T, Liu H-W, Postman J, Li R (2018) Molecular characterization of a novel rhabdovirus infecting blackcurrant identified by high-throughput sequencing. Arch Virol 163:1363–1366
CAS Google Scholar - Wu Z, Du J, Lu L, Yang L, Dong J, Sun L, Zhu Y, Liu Q, Jin Q (2018) Detection of Hantaviruses and Arenaviruzses [sic] in three-toed jerboas from the Inner Mongolia Autonomous Region, China. Emerg Microbes Infect 7:35
CAS PubMed Central Google Scholar - Wu Z, Lu L, Du J, Yang L, Ren X, Liu B, Jiang J, Yang J, Dong J, Sun L, Zhu Y, Li Y, Zheng D, Zhang C, Su H, Zheng Y, Zhou H, Zhu G, Li H, Chmura A, Yang F, Daszak P, Wang J, Liu Q, Jin Q (2018) Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 6:178
PubMed Central Google Scholar - Xu C, Sun X, Taylor A, Jiao C, Xu Y, Cai X, Wang X, Ge C, Pan G, Wang Q, Fei Z, Wang Q (2017) Diversity, distribution, and evolution of tomato viruses in China uncovered by small RNA sequencing. J Virol 91:e00173-17
PubMed Central Google Scholar - Xu FL, Liu DY, Nunes MRT, Da Rosa A, Tesh RB, Xiao SY (2007) Antigenic and genetic relationships among Rift Valley fever virus and other selected members of the genus Phlebovirus (Bunyaviridae). Am J Trop Med Hyg 76:1194–1200
CAS Google Scholar - Xu Y, Lou S-G, Li XL, Zheng Y-X, Wang W-C, Liu Y-T (2013) The complete S RNA and M RNA nucleotide sequences of a hippeastrum chlorotic ringspot virus (HCRV) isolate from Hymenocallis littoralis (Jacq.) Salisb in China. Arch Virol 158:2597–2601
CAS Google Scholar - Yadav PD, Nyayanit DA, Shete AM, Jain S, Majumdar TP, Chaubal GY, Shil P, Kore PM, Mourya DT (2019) Complete genome sequencing of Kaisodi virus isolated from ticks in India belonging to Phlebovirus genus, family Phenuiviridae. Ticks Tick Borne Dis 10:23–33
CAS Google Scholar - Yang X-L, Zhang Y-Z, Jiang R-D, Guo H, Zhang W, Li B, Wang N, Wang L, Waruhiu C, Zhou J-H, Li S-Y, Daszak P, Wang L-F, Shi Z-L (2017) Genetically diverse filoviruses in Rousettus and Eonycteris spp. bats, China, 2009 and 2015. Emerg Infect Dis 23:482–486
PubMed Central Google Scholar - Yang X-L, Tan CW, Anderson DE, Jiang R-D, Li B, Zhang W, Zhu Y, Lim XF, Zhou P, Liu X-L, Guan W, Zhang L, Li S-Y, Zhang Y-Z, Wang L-F, Shi Z-L (2019) Characterization of a filovirus (Měnglà virus) from Rousettus bats in China. Nat Microbiol 4:390–395
CAS Google Scholar - Yang X, Huang J, Liu C, Chen B, Zhang T, Zhou G (2016) Rice stripe mosaic virus, a novel cytorhabdovirus infecting rice via leafhopper transmission. Front Microbiol 7:2140
Google Scholar - Zhao G, Krishnamurthy S, Cai Z, Popov VL, Travassos da Rosa AP, Guzman H, Cao S, Virgin HW, Tesh RB, Wang D (2013) Identification of novel viruses using VirusHunter - an automated data analysis pipeline. PLoS One 8:e78470
CAS PubMed Central Google Scholar - Бyтeнкo AM, Гpoмaшeвcкий BЛ, Львoв ДК, Пoпoв BФ (1979) Bиpyc Киceмaйo—пpeдcтaвитeль aнтигeннoй гpyппы Бxaнджa. Boпp Bиpycoл:661–5
- Львoв ДК, Aльxoвcкий CB, Щeлкaнoв MЮ, Щeтинин AM, Дepябин ПГ, Apиcтoвa BA, Гитeльмaн AК, Caмoxвaлoв EИ, Бoтикoв AГ (2014) Гeнeтичecкaя xapaктepиcтикa виpycoв Caxaлин (SAKV - Sakhalin virus), Пapaмyшиp (PMRV - Paramushir virus) (Bunyaviridae, Nairovirus, гpyппa Caxaлин) и Pyкyтaмa (RUKV - Rukutama virus) (Bunyaviridae, Phlebovirus, гpyппa Укyниeми), изoлиpoвaнныx oт oблигaтныx пapaзитoв кoлoниaльныx мopcкиx птиц - клeщeй Ixodes (Ceratixodes) uriae, White 1852 и I. signatus Birulya, 1895 в бacceйнax Oxoтcкoгo и Бepингoвa мopeй. Boпp Bиpycoл 59:11–17
Google Scholar - Львoв ДК, Aльxoвcкий CB, Щeлкaнoв MЮ, Щeтинин AM, Дepябин ПГ, Гитeльмaн AК, Caмoxвaлoв EИ, Бoтикoв AГ (2014) Гeнeтичecкaя xapaктepиcтикa штaммoв виpyca Зaлив Tepпeния (ZTV - Zaliv Terpeniya virus) (Bunyaviridae, Phlebovirus, aнтигeнный кoмплeкc Укyниeми), изoлиpoвaннoгo в выcoкиx шиpoтax Ceвepнoй Eвpaзии из oблигaтныx эктoпapaзитoв чиcтикoвыx птиц (Alcidae Leach, 1820)—клeщeй Ixodes (Ceratixodes) uriae White, 1852 и oт кoмapoв Culex modestus Ficalbi, 1889 в cyбтpoпикax Зaкaвкaзья. Boпp Bиpycoл 59:12–18
Google Scholar
Acknowledgements
We thank W. Ian Lipkin and Rafal Tokarz (Columbia University Irving Medical Center, New York, New York, USA) for providing/approving new names for “blacklegged tick phleboviruses 1 and 3” and Edward Holmes (University of Sydney, Australia) for providing/approving a new name for “Norway phlebovirus”. Many authors are current members of 2017-2020 International Committee on Taxonomy of Viruses (ICTV) Study Groups: Arenaviridae (Jens H. Kuhn, Michael J. Buchmeier, Rémi N. Charrel, J. Christopher S. Clegg, Juan Carlos de la Torre, Jean-Paul J. Gonzalez, Stephan Günther, Mark D. Stenglein, Jussi Hepojoki, Manuela Sironi, Igor S. Lukashevich, Sheli R. Radoshitzky, Víctor Romanowski, Maria S. Salvato), Artoviridae (Jens H. Kuhn, Ralf G. Dietzgen, Dàohóng Jiāng, Nikos Vasilakis), Aspiviridae (John V. da Graça, Elena Dal Bó, Selma Gago-Zachert, María Laura García, John Hammond, Tomohide Natsuaki, José A. Navarro, Vicente Pallás, Carina A. Reyes, Gabriel Robles Luna, Takahide Sasaya, Ioannis Tzanetakis, Anna Maria Vaira, Martin Verbeek), Bornaviridae (Jens H. Kuhn, Thomas Briese, Ralf Dürrwald, Masayuki Horie, Timothy H. Hyndman, Norbert Nowotny, Susan Payne, Dennis Rubbenstroth, Mark D. Stenglein, Keizō Tomonaga), Bunyavirales (Jens H. Kuhn, Scott Adkins, Juan Carlos de la Torre, Sandra Junglen, Amy J. Lambert, Piet Maes, Marco Marklewitz, Gustavo Palacios, Takahide Sasaya, Yong-Zhen Zhang), Filoviridae (Jens H. Kuhn, Gaya K. Amarasinghe, Christopher Basler, Sina Bavari, Alexander Bukreyev, Kartik Chandran, Ian Crozier, Olga Dolnik, John M. Dye, Pierre B. H. Formenty, Anthony Griffiths, Roger Hewson, Gary Kobinger, Eric M. Leroy, Elke Mühlberger, Sergey V. Netesov, Gustavo Palacios, Bernadett Pályi, Janusz T. Pawęska, Sophie Smither, Ayato Takada, Jonathan S. Towner, Victoria Wahl), Fimoviridae (Michele Digiaro, Toufic Elbeaino, Giovanni P. Martelli, Nicole Mielke-Ehret, Hans-Peter Mühlbach), Hantaviridae (Steven Bradfute, Charles H. Calisher, Boris Klempa, Jonas Klingström, Lies Laenen, Piet Maes, Jin-Won Song, Yong-Zhen Zhang), Jingchuvirales (Nicholas Di Paola), Monjiviricetes (Jens H. Kuhn, Ralf G. Dietzgen, W. Paul Duprex, Dàohóng Jiāng, Piet Maes, Janusz T. Pawęska, Bertus K. Rima, Dennis Rubbenstroth, Peter J. Walker, Yong-Zhen Zhang), Mymonaviridae (María A. Ayllón, Dàohóng Jiāng, Shin-Yi L. Marzano), Nairoviridae (Jens H. Kuhn, Sergey V. Alkhovsky, Tatjana Avšič-Županc, Dennis A. Bente, Éric Bergeron, Felicity Burt, Nicholas Di Paola, Koray Ergünay, Aura R. Garrison, Roger Hewson, Ali Mirazimi, Gustavo Palacios, Anna Papa, Amadou Alpha Sall, Jessica R. Spengler), Negarnaviricota (Jens H. Kuhn, Eugene V. Koonin, Mart Krupovic, Yuri I. Wolf), Nyamiviridae (Jens H. Kuhn, Ralf G. Dietzgen, Dàohóng Jiāng, Nikos Vasilakis), Orthomyxoviridae (Justin Bahl, Inmaculada Casas, Adolfo García-Sastre, Seiji Hongo, Sergio H. Marshall, John W. McCauley, Gabriele Neumann, Colin R. Parrish, Daniel R. Pérez, Jonathan A. Runstadler, Martin Schwemmle), Paramyxoviridae (Anne Balkema-Buschmann, William G. Dundon, W. Paul Duprex, Andrew J. Easton, Ron Fouchier, Gael Kurath, Benhur Lee, Bertus K. Rima, Paul Rota, Lin-Fa Wang, Robobert A. Lamb), Peribunyaviridae (Scott Adkins, Sergey V. Alkhovsky, Martin Beer, Carol D. Blair, Charles H. Calisher, Michael A. Drebot, Holly R. Hughes, Amy J. Lambert, William Marciel de Souza, Marco Marklewitz, Márcio R. T. Nunes, Xiǎohóng Shí), Phasmaviridae (Matthew J. Ballinger, Roy A. Hall, Sandra Junglen, Stanley L. Langevin, Alex Pauvolid-Corrêa), Phenuiviridae (Thomas Briese, Rémi N. Charrel, Xavier De Lamballerie, Hideki Ebihara, George Fú Gāo, Martin H. Groschup, Márcio R. T. Nunes, Gustavo Palacios, Takahide Sasaya, Jin-Won Song), Pneumoviridae (Paul A. Brown, Ursula J. Buchholz, Rik L. de Swart, Jan Felix Drexler, W. Paul Duprex, Andrew J. Easton, Jiànróng Lǐ, Kirsten Spann, Natalie J. Thornburg, Bernadette van den Hoogen, John V. Williams), Rhabdoviridae (Kim R. Blasdell, Rachel Breyta, Ralf G. Dietzgen, Anthony R. Fooks, Juliana Freitas-Astúa, Hideki Kondō, Gael Kurath, Ivan V. Kuzmin, David M. Stone, Robert B. Tesh, Noël Tordo, Nikos Vasilakis, Peter J. Walker, Anna E. Whitfield), Sunviridae (Timothy H. Hyndman, Gael Kurath), Tenuivirus (Il-Ryong Choi, Gilda B. Jonson, Takahide Sasaya, Yukio Shirako, Tàiyún Wèi, Xueping Zhou), and Tospoviridae (Scott Adkins, Amy J. Lambert, Rayapati Naidu, Renato O. Resende, Massimo Turina, Anna E. Whitfield); or are ICTV Executive Committee Members: the 2017–2020 ICTV Chair of the Fungal and Protist Viruses Subcommittee (Peter Simmonds), the 2018–2020 ICTV Proposal Secretary (Peter J. Walker), the 2017–2020 ICTV Chair of the Plant Viruses Subcommittee (F. Murilo Zerbini), the 2017–2020 ICTV Chair of the Animal dsRNA and ssRNA- Viruses Subcommittee (Jens H. Kuhn), and 2017–2020 Elected Members (Sead Sabanadzovic, Arvind Varsani). We would like to thank Anya Crane (NIH/NIAID/DCR/IRF-Frederick) for critically editing the manuscript.
Author information
Author notes
- Giovanni P. Martelli: Deceased.
Authors and Affiliations
- Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Fort Detrick, Frederick, MD, USA
Jens H. Kuhn - United States Department of Agriculture, Agricultural Research Service, US Horticultural Research Laboratory, Fort Pierce, FL, USA
Scott Adkins - Dipartimento di Agraria, Università degli Studi di Napoli Federico II, Portici, Italy
Daniela Alioto & Maria Minutolo - D.I. Ivanovsky Institute of Virology of N.F. Gamaleya National Center on Epidemiology and Microbiology of Ministry of Health of Russian Federation, Moscow, Russia
Sergey V. Alkhovsky - Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO, USA
Gaya K. Amarasinghe - Mailman School of Public Health, Columbia University, New York, NY, USA
Simon J. Anthony - EcoHealth Alliance, New York, NY, USA
Simon J. Anthony - Ljubljana Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia
Tatjana Avšič-Županc - Centro de Biotecnología y Genómica de Plantas, Universidad Politécnica de Madrid-Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria, Campus de Montegancedo, Pozuelo de Alarcón, Madrid, Spain
María A. Ayllón - Departamento de Biotecnología-Biología Vegetal, Escuela Técnica Superior de Ingeniería Agronómica, Alimentaria y de Biosistemas, Universidad Politécnica de Madrid, Madrid, Spain
María A. Ayllón - Department of Infectious Diseases, Department of Epidemiology and Biostatistics, Institute of Bioinformatics, Center for Ecology of Infectious Diseases, University of Georgia, Athens, GA, USA
Justin Bahl - Friedrich-Loeffler-Institut, Federal Research Institute for Animal Health, Institute of Novel and Emerging Infectious Diseases, Greifswald-Insel Riems, Germany
Anne Balkema-Buschmann & Martin H. Groschup - Department of Biological Sciences, Mississippi State University, Mississippi State, MS, USA
Matthew J. Ballinger - Department of Botany and Zoology, Masaryk University, Brno, Czech Republic
Tomáš Bartonička - Center for Microbial Pathogenesis, Institute for Biomedical Sciences, Georgia State University, Atlanta, GA, USA
Christopher Basler - Edge BioInnovation Consulting and Mgt, Frederick, MD, USA
Sina Bavari - Institute of Diagnostic Virology, Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany
Martin Beer, Leonie F. Forth, Florian Pfaff & Dennis Rubbenstroth - Galveston National Laboratory, The University of Texas, Medical Branch at Galveston, Galveston, TX, USA
Dennis A. Bente & Alexander Bukreyev - Viral Special Pathogens Branch, Division of High-Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA, USA
Éric Bergeron, Jessica R. Spengler & Jonathan S. Towner - School of Veterinary Medicine, One Health Institute, University of California, Davis, CA, USA
Brian H. Bird - Department of Microbiology, Immunology and Pathology, Colorado State University, Fort Collins, CO, USA
Carol Blair - Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australian Centre for Disease Preparedness, Geelong, VIC, Australia
Kim R. Blasdell - University of New Mexico Health Sciences Center, Albuquerque, NM, USA
Steven B. Bradfute - US Geological Survey, Western Fisheries Research Center, Seattle, WA, USA
Rachel Breyta - Department of Epidemiology, Mailman School of Public Health, Center for Infection and Immunity, Columbia University, New York, NY, USA
Thomas Briese - Laboratory of Ploufragan-Plouzané-Niort, French Agency for Food, Environmental and Occupational Heath Safety ANSES, Ploufragan, France
Paul A. Brown - RNA Viruses Section, Laboratory of Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA
Ursula J. Buchholz - Department of Molecular Biology and Biochemistry, University of California, Irvine, CA, USA
Michael J. Buchmeier - Department of Pathology, The University of Texas Medical Branch, Galveston, TX, USA
Alexander Bukreyev, Robert B. Tesh & Nikos Vasilakis - Division of Virology, National Health Laboratory Service, University of the Free State, Bloemfontein, Republic of South Africa
Felicity Burt - Department of Plant Protection, Faculty of Agriculture, Kahramanmaras Sütçü Imam University, Avsar Campus, 46060, Kahramanmaras, Turkey
Nihal Buzkan - Colorado State University, Fort Collins, CO, USA
Charles H. Calisher - National Citrus Engineering Research Center, Citrus Research Institute, Southwest University, Chongqing, 400712, China
Mengji Cao - Academy of Agricultural Sciences, Southwest University, Chongqing, 400715, China
Mengji Cao - Respiratory Virus and Influenza Unit, National Microbiology Center, Instituto de Salud Carlos III, Madrid, Spain
Inmaculada Casas - Virology and Pathogenesis Group, National Infection Service, Public Health England, Porton Down, UK
John Chamberlain - Department of Microbiology and Immunology, Albert Einstein College of Medicine, Bronx, NY, USA
Kartik Chandran - Unité des Virus Emergents (Aix-Marseille Univ-IRD 190-Inserm 1207-IHU Méditerranée Infection), Marseille, France
Rémi N. Charrel & Xavier de Lamballerie - Guangdong Province Key Laboratory of Microbial Signals and Disease Control, College of Agriculture, South China Agricultural University, Guangdong, China
Biao Chen, Xin Yang, Tong Zhang & Guohui Zhou - Istituto per la Protezione Sostenibile delle Piante-Consiglio Nazionale delle ricerche (Institute for Sustainable Plant Protection-National Research Council), Bari, Italy
Michela Chiumenti, Francesco Di Serio, Angelantonio Minafra & Beatriz Navarro - Plant Breeding Genetics and Biotechnology Division, International Rice Research Institute, Los Baños, Philippines
Il-Ryong Choi - Les Mandinaux, Le Grand Madieu, France
J. Christopher S. Clegg - Clinical Monitoring Research Program Directorate, Frederick National Laboratory for Cancer Research, Frederick, MD, USA
Ian Crozier - Texas A&M University-Kingsville Citrus Center, Weslaco, TX, USA
John V. da Graça - CIDEFI. Facultad de Ciencias Agrarias y Forestales, Universidad de La Plata, La Plata, Argentina
Elena Dal Bó - Laboratório de Biologia Computacional e Sistemas, Fundação Oswaldo Cruz-Fiocruz, Instituto Oswaldo Cruz, Rio de Janeiro, RJ, Brasil
Alberto M. R. Dávila & Rodrigo Jardim - Department of Immunology and Microbiology IMM-6, The Scripps Research Institute, La Jolla, CA, USA
Juan Carlos de la Torre - Department Viroscience, Erasmus MC University Medical Centre Rotterdam, Rotterdam, The Netherlands
Rik L. de Swart & Ron A. M. Fouchier - Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR, 97331, USA
Patrick L. Di Bello - United States Army Medical Research Institute of Infectious Diseases, Fort Detrick, Frederick, MD, USA
Nicholas Di Paola, John M. Dye, Aura R. Garrison, Gustavo Palacios & Sheli R. Radoshitzky - Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, St. Lucia, QLD, Australia
Ralf G. Dietzgen - CIHEAM, Istituto Agronomico Mediterraneo di Bari, Valenzano, Italy
Michele Digiaro - Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR, USA
Valerian V. Dolja - Institute of Virology, Philipps University Marburg, Marburg, Germany
Olga Dolnik - Zoonotic Diseases and Special Pathogens, National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, MB, Canada
Michael A. Drebot - Institute of Virology, Charité-Universitätsmedizin Berlin, corporate member of Free University Berlin, Humboldt-University Berlin, and Berlin Institute of Health, Berlin, Germany
Jan Felix Drexler, Sandra Junglen & Marco Marklewitz - Robert Koch Institut, Berlin, Germany
Ralf Dürrwald - Veterinary Research Institute, Brno, Czech Republic
Lucie Dufkova, Daniel Ruzek, Jiří Salát, Jana Širmarová & Petra Straková - Animal Production and Health Laboratory, Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture, Department of Nuclear Sciences and Applications, International Atomic Energy Agency, Vienna, Austria
William G. Dundon - School of Medicine, University of Pittsburgh, Pittsburgh, PA, USA
W. Paul Duprex & John V. Williams - School of Life Sciences, University of Warwick, Coventry, UK
Andrew J. Easton - Department of Molecular Medicine, Mayo Clinic, Rochester, MN, USA
Hideki Ebihara - Istituto Agronomico Mediterraneo di Bari, Valenzano, Italy
Toufic Elbeaino - Virology Unit, Department of Medical Microbiology, Faculty of Medicine, Hacettepe University, Ankara, Turkey
Koray Ergünay - Animal and Plant Health Agency, Weybridge, Surrey, UK
Anthony R. Fooks - World Health Organization, Geneva, Switzerland
Pierre B. H. Formenty - Embrapa Cassava and Fruits, Cruz das Almas, Bahia, Brazil
Juliana Freitas-Astúa - Institute of Biochemistry and Biotechnology, Martin Luther University Halle-Wittenberg, Halle/Saale, Germany
Selma Gago-Zachert - Department of Molecular Signal Processing, Leibniz Institute of Plant Biochemistry, Halle/Saale, Germany
Selma Gago-Zachert - National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
George Fú Gāo & Dexin Li - Instituto de Biotecnología y Biología Molecular, Facultad de Ciencias Exactas, CONICET UNLP, La Plata, Argentina
María Laura García - Icahn School of Medicine at Mount Sinai, New York, NY, USA
Adolfo García-Sastre - Metabiota, Inc. Sierra Leone, Freetown, Sierra Leone
Aiah Gbakima - One Health Institute, Karen C. Drayer Wildlife Health Center, School of Veterinary Medicine, University of California, Davis, CA, USA
Tracey Goldstein - Department of Microbiology and Immunology, Division of Biomedical Graduate Research Organization, School of Medicine, Georgetown University, Washington, DC, 20057, USA
Jean-Paul J. Gonzalez - Centaurus Biotechnologies, CTP, Manassas, VA, USA
Jean-Paul J. Gonzalez - Department of Microbiology and National Emerging Infectious Diseases Laboratories, Boston University School of Medicine, Boston, MA, USA
Anthony Griffiths & Elke Mühlberger - Department of Virology, Bernhard-Nocht Institute for Tropical Medicine, WHO Collaborating Centre for Arboviruses and Hemorrhagic Fever Reference and Research, Hamburg, Germany
Stephan Günther - Australian Infectious Diseases Research Centre, School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Australia
Roy A. Hall - United States Department of Agriculture, Agricultural Research Service, USNA, Floral and Nursery Plants Research Unit, Beltsville, MD, USA
John Hammond - Department of Agricultural Botany, Faculty of Agriculture, Fayoum University, Fayoum, Egypt
Mohamed Hassan - Department of Virology, University of Helsinki, Medicum, Helsinki, Finland
Jussi Hepojoki, Satu Hepojoki & Teemu Smura - Vetsuisse Faculty, Institute of Veterinary Pathology, University of Zurich, Zurich, Switzerland
Jussi Hepojoki - Mobidiag Ltd, Espoo, Finland
Satu Hepojoki - Institute of Veterinary Pathology, University of Zuerich, Zurich, Switzerland
Udo Hetzel - Public Health England, Porton Down, Salisbury, Wiltshire, UK
Roger Hewson - Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany
Bernd Hoffmann & Dirk Höper - Department of Infectious Diseases, Faculty of Medicine, Yamagata University, Yamagata, Japan
Seiji Hongo - Hakubi Center for Advanced Research, Kyoto University, Kyoto, Japan
Masayuki Horie - School of Veterinary Medicine, Murdoch University, Murdoch, WA, Australia
Timothy H. Hyndman - Ministry of Health and Sanitation, Freetown, Sierra Leone
Amara Jambai - State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, Hubei, China
Dàohóng Jiāng - Ministry of Health Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
Qi Jin - Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Hangzhou, China
Qi Jin - Department of Agricultural Biotechnology, Center for Fungal Pathogenesis, College of Agriculture and Life Sciences, Seoul National University, Seoul, South Korea
Gilda B. Jonson & Piet Maes - Republic Of Turkey Ministry Of Agriculture And Forestry, Pistachio Research Institute, Gaziantep, Turkey
Serpil Karadağ - United States Department of Agriculture, Agricultural Research Service, Horticulture Crops Research Unit, Corvallis, OR, USA
Karen E. Keller - Institute of Virology, Biomedical Research Center, Slovak Academy of Sciences, Bratislava, Slovakia
Boris Klempa - Department of Medicine Huddinge, Center for Infectious Medicine, Karolinska Institutet, Karolinska University Hospital, Stockholm, Sweden
Jonas Klingström - Department of Microbiology, Immunology and Infectious Diseases, Université Laval, Quebec City, Canada
Gary Kobinger - Institute of Plant Science and Resources, Okayama University, Kurashiki, Japan
Hideki Kondō - National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD, USA
Eugene V. Koonin, Yuri I. Wolf & Natalya Yutin - Archaeal Virology Unit, Institut Pasteur, Paris, France
Mart Krupovic - US Geological Survey Western Fisheries Research Center, Seattle, WA, USA
Gael Kurath - US Department of Agriculture, Animal and Plant Health Inspection, National Veterinary Services Laboratories, Diagnostic Virology Laboratory, Ames, USA
Ivan V. Kuzmin - Zoonotic Infectious Diseases Unit, KU Leuven, Rega Institute, Leuven, Belgium
Lies Laenen - Department of Laboratory Medicine, University Hospitals Leuven, Leuven, Belgium
Lies Laenen - Department of Molecular Biosciences, Northwestern University, Evanston, IL, USA
Robert A. Lamb - Howard Hughes Medical Institute, Northwestern University, Evanston, IL, USA
Robert A. Lamb - Department of Microbiology, University of Washington, Washington, USA
Stanley L. Langevin - Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, NY, USA
Benhur Lee - MIVEGEC (IRD-CNRS-Montpellier university) Unit, French National Research Institute for Sustainable Development (IRD), Montpellier, France
Eric M. Leroy - Department of Veterinary Biosciences, College of Veterinary Medicine, The Ohio State University, Columbus, OH, USA
Jiànróng Lǐ - Key Laboratory for Medical Virology, NHFPC, National Institute for Viral Disease Control and Prevention, Beijing, China
Mifang Liang - State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Wénwén Liú, Yàn Liú, Hui Wang, Xifeng Wang & Xueping Zhou - Department of Pharmacology and Toxicology, School of Medicine, The Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases, University of Louisville, Louisville, KY, USA
Igor S. Lukashevich - Virology Research Center, University of São Paulo, Ribeirão Preto, Brazil
William Marciel de Souza - German Center for Infection Research (DZIF), Berlin, Germany
Sandra Junglen & Marco Marklewitz - Pontificia Universidad Católica de Valparaíso, Campus Curauma, Valparaíso, Chile
Sergio H. Marshall - Department of Plant, Soil and Food Sciences, University “Aldo Moro”, Bari, Italy
Giovanni P. Martelli - United States Department of Agriculture, Horticultural Crops Research Unit, Corvallis, OR, USA
Robert R. Martin - Department of Biology and Microbiology, Department of Plant Sciences, South Dakota State University, Brookings, SD, USA
Shin-Yi L. Marzano - Gembloux Agro-Bio Tech, TERRA, Plant Pathology Laboratory, Liège University, Liège, Belgium
Sébastien Massart - Worldwide Influenza Centre, Francis Crick Institute, London, UK
John W. McCauley - Biocentre Klein Flottbek, University of Hamburg, Hamburg, Germany
Nicole Mielke-Ehret & Hans-Peter Mühlbach - Folkhalsomyndigheten, Stockholm, Sweden
Ali Mirazimi - Department of Plant Pathology, Irrigated Agricultural Research and Extension Center, Washington State University, Prosser, WA, USA
Rayapati Naidu - School of Agriculture, Utsunomiya University, Utsunomiya, Tochigi, Japan
Tomohide Natsuaki - Instituto de Biología Molecular y Celular de Plantas, Universitat Politècnica de València-Consejo Superior de Investigaciones Científicas, Valencia, Spain
José A. Navarro - Novosibirsk State University, Novosibirsk, Novosibirsk Oblast, Russia
Sergey V. Netesov - Department of Pathobiological Sciences, Influenza Research Institute, University of Wisconsin-Madison, Madison, USA
Gabriele Neumann - Institute of Virology, University of Veterinary Medicine Vienna, Vienna, Austria
Norbert Nowotny - College of Medicine, Mohammed Bin Rashid University of Medicine and Health Sciences, Dubai, United Arab Emirates
Norbert Nowotny - Evandro Chagas Institute, Ministry of Health, Pará, Brazil
Márcio R. T. Nunes - Fish Disease Research Group, Department of Biological Sciences, University of Bergen, Bergen, Norway
Are Nylund & Arnfinn L. Økland - Instituto de Biología Molecular y Celular de Plantas (IBMCP), Consejo Superior de Investigaciones Cientificas-Universidad Politécnica de Valencia, Valencia, Spain
Vicente Pallas - National Biosafety Laboratory, National Public Health Center, Budapest, Hungary
Bernadett Pályi - National Reference Centre for Arboviruses and Haemorrhagic Fever Viruses, Department of Microbiology, Medical School, Aristotle University of Thessaloniki, Thessaloníki, Greece
Anna Papa - College of Veterinary Medicine, Baker Institute for Animal Health, Cornell University, Ithaca, NY, USA
Colin R. Parrish - Department of Veterinary Integrated Biosciences and Department of Entomology, Texas A&M University, College Station, USA
Alex Pauvolid-Corrêa - Center for Emerging Zoonotic and Parasitic Diseases, National Institute for Communicable Diseases of the National Health Laboratory Service, Sandringham-Johannesburg, Gauteng, South Africa
Janusz T. Pawęska - Department of Veterinary Pathobiology, College of Veterinary Medicine and Biomedical Sciences, Texas A&M University, College Station, TX, USA
Susan Payne - Department of Population Health, College of Veterinary Medicine, University of Georgia, Athens, GA, USA
Daniel R. Pérez - Institute of Microbiology, University of Veterinary and Animal Sciences, Lahore, Pakistan
Aziz-ul Rahman & Muhammad Z. Shabbir - Laboratório de Biologia Molecular Aplicada, Instituto Biológico, São Paulo, SP, Brazil
Pedro L. Ramos-González - Departamento de Biologia Celular, Universidade de Brasília, Brasília, Brazil
Renato O. Resende - Instituto de Biotecnología y Biología Molecular, CCT-La Plata, CONICET-UNLP, La Plata, Buenos Aires, Argentina
Carina A. Reyes & Gabriel Robles Luna - Centre for Experimental Medicine, School of Medicine, Dentistry and Biomedical Sciences, The Queen’s University of Belfast, Belfast, Northern Ireland, UK
Bertus K. Rima - Instituto de Biotecnología y Biología Molecular, Centro Cientifico Technológico-La Plata, Consejo Nacional de Investigaciones Científico Tecnológico-Universidad Nacional de La Plata, La Plata, Argentina
Víctor Romanowski - National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, Atlanta, GA, USA
Paul Rota - Department of Infectious Disease and Global Health, Tufts University Cummings School of Veterinary Medicine, 200 Westboro Road, North Grafton, MA, 01536, USA
Jonathan A. Runstadler - Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, Branisovska 31, 37005, Ceske Budejovice, Czech Republic
Daniel Ruzek & Jiří Salát - Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology, Mississippi State University, Mississippi State, MS, USA
Sead Sabanadzovic - Institut Pasteur de Dakar, Dakar, Senegal
Amadou Alpha Sall - Institute of Human Virology, University of Maryland School of Medicine, Baltimore, MD, USA
Maria S. Salvato - Department of Forestry Engineering, Faculty of Forestry, Karabuk University (UNIKA), Karabük, Turkey
Kamil Sarpkaya - Western Region Agricultural Research Center, National Agriculture and Food Research Organization, Fukuyama, Japan
Takahide Sasaya - Faculty of Medicine, University Medical Center-University Freiburg, Freiburg, Germany
Martin Schwemmle - MRC-University of Glasgow Centre for Virus Research, Glasgow, Scotland, UK
Xiǎohóng Shí - CAS Key Laboratory of Special Pathogens, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, Wuhan, Hubei, People’s Republic of China
Zhènglì Shí & Xīnglóu Yáng - Asian Center for Bioresources and Environmental Sciences, University of Tokyo, Tokyo, Japan
Yukio Shirako - Nuffield Department of Medicine, University of Oxford, Oxford, UK
Peter Simmonds - Bioinformatics Unit, Scientific Institute IRCCS “E. Medea”, Bosisio Parini, Italy
Manuela Sironi - CBR Division, Dstl, Porton Down, Salisbury, Wiltshire, UK
Sophie Smither - Department of Microbiology, College of Medicine, Korea University, Seoul, Republic of Korea
Jin-Won Song - School of Biomedical Sciences, Faculty of Health, Queensland University of Technology, Brisbane, QLD, Australia
Kirsten M. Spann - Department of Microbiology, Immunology, and Pathology, College of Veterinary Medicine and Biomedical Sciences, Colorado State University, Fort Collins, CO, USA
Mark D. Stenglein - Centre for Environment, Fisheries and Aquaculture Science, Weymouth, Dorset, UK
David M. Stone - Division of Global Epidemiology, Research Center for Zoonosis Control, Hokkaido University, Sapporo, Japan
Ayato Takada - Centers for Disease Control and Prevention, Atlanta, GA, USA
Natalie J. Thornburg - Institute for Frontier Life and Medical Sciences (inFront), Kyoto University, Kyoto, Japan
Keizō Tomonaga - Institut Pasteur, Unité des Stratégies Antivirales, WHO Collaborative Centre for Viral Haemorrhagic Fevers and Arboviruses, OIE Reference Laboratory for RVFV and CCHFV, Paris, France
Noël Tordo - Institut Pasteur de Guinée, Conakry, Guinea
Noël Tordo - Institute for Sustainable Plant Protection, National Research Council of Italy (CNR), Strada delle Cacce 73, 10135, Turin, Italy
Massimo Turina - Division of Agriculture, Department of Entomology and Plant Pathology, University of Arkansas System, Fayetteville, AR, 72701, USA
Ioannis Tzanetakis - Friedrich-Loeffler-Institut, Federal Research Institute for Animal Health, Institute of Novel and Emerging Infectious Diseases, Südufer 10, 17493, Greifswald-Insel Riems, Germany
Rainer G. Ulrich - German Center of Infection Research (DZIF), Partner site Hamburg-Lübeck-Borstel-Insel Riems, Greifswald-Insel Riems, Germany
Rainer G. Ulrich - Institute for Sustainable Plant Protection, National Research Council of Italy (IPSP-CNR), 73 Strada delle Cacce, 10135, Turin, Italy
Anna Maria Vaira - Department of Viroscience, Erasmus MC University Medical Centre Rotterdam, Rotterdam, The Netherlands
Bernadette van den Hoogen - The Biodesign Center for Fundamental and Applied Microbiomics, Center for Evolution and Medicine School of Life Sciences, Arizona State University, Tempe, AZ, USA
Arvind Varsani - Structural Biology Research Unit, Department of Clinical Laboratory Sciences, University of Cape Town, Observatory, Cape Town, South Africa
Arvind Varsani - Wageningen University and Research, Biointeractions and Plant Health, Wageningen, The Netherlands
Martin Verbeek - National Biodefense Analysis and Countermeasures Center, Fort Detrick, Frederick, MD, USA
Victoria Wahl - NHC Key Laboratory of Systems Biology of Pathogens and Christophe Mérieux Laboratory, IPB-Fondation Mérieux, Institute of Pathogen Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
Jianwei Wang - Programme in Emerging Infectious Diseases, Duke-NUS Medical School, Singapore, Singapore
Lin-Fa Wang - Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China
Tàiyún Wèi - Mailman School of Public Health, Center for Infection and Immunity, Columbia University, New York, USA
Heather Wells - Department of Entomology and Plant Pathology, North Carolina State University, Raleigh, NC, USA
Anna E. Whitfield - MOH Key Laboratory of Systems Biology of Pathogens, IPB, CAMS, Beijing, China
Zhìqiáng Wú - Wuhan University School of Health Sciences, Wuhan, China
Xuejie Yu - Departamento de Fitopatologia, Instituto de Biotecnologia Aplicada à Agropecuária, Universidade Federal de Viçosa, Viçosa, Minas Gerais, Brazil
F. Murilo Zerbini - National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing, China
Yong-Zhen Zhang - Shanghai Public Health Clinical Center, Institutes of Biomedical Sciences, Fudan University, Shanghai, China
Yong-Zhen Zhang - Centers for Disease Control and Prevention, Fort Collins, CO, USA
Holly R. Hughes & Amy J. Lambert - Laboratório de Hantaviroses e Rickettsioses, Fundação Oswaldo Cruz-Fiocruz, Instituto Oswaldo Cruz, Rio de Janeiro, RJ, Brasil
Jorlan Fernandes, Alexandro Guterres, Elba R. S. Lemos & Renata C. Oliveira - School of Biological Sciences, University of Queensland, St. Lucia, Queensland, Australia
Peter J. Walker
Authors
- Jens H. Kuhn
- Scott Adkins
- Daniela Alioto
- Sergey V. Alkhovsky
- Gaya K. Amarasinghe
- Simon J. Anthony
- Tatjana Avšič-Županc
- María A. Ayllón
- Justin Bahl
- Anne Balkema-Buschmann
- Matthew J. Ballinger
- Tomáš Bartonička
- Christopher Basler
- Sina Bavari
- Martin Beer
- Dennis A. Bente
- Éric Bergeron
- Brian H. Bird
- Carol Blair
- Kim R. Blasdell
- Steven B. Bradfute
- Rachel Breyta
- Thomas Briese
- Paul A. Brown
- Ursula J. Buchholz
- Michael J. Buchmeier
- Alexander Bukreyev
- Felicity Burt
- Nihal Buzkan
- Charles H. Calisher
- Mengji Cao
- Inmaculada Casas
- John Chamberlain
- Kartik Chandran
- Rémi N. Charrel
- Biao Chen
- Michela Chiumenti
- Il-Ryong Choi
- J. Christopher S. Clegg
- Ian Crozier
- John V. da Graça
- Elena Dal Bó
- Alberto M. R. Dávila
- Juan Carlos de la Torre
- Xavier de Lamballerie
- Rik L. de Swart
- Patrick L. Di Bello
- Nicholas Di Paola
- Francesco Di Serio
- Ralf G. Dietzgen
- Michele Digiaro
- Valerian V. Dolja
- Olga Dolnik
- Michael A. Drebot
- Jan Felix Drexler
- Ralf Dürrwald
- Lucie Dufkova
- William G. Dundon
- W. Paul Duprex
- John M. Dye
- Andrew J. Easton
- Hideki Ebihara
- Toufic Elbeaino
- Koray Ergünay
- Jorlan Fernandes
- Anthony R. Fooks
- Pierre B. H. Formenty
- Leonie F. Forth
- Ron A. M. Fouchier
- Juliana Freitas-Astúa
- Selma Gago-Zachert
- George Fú Gāo
- María Laura García
- Adolfo García-Sastre
- Aura R. Garrison
- Aiah Gbakima
- Tracey Goldstein
- Jean-Paul J. Gonzalez
- Anthony Griffiths
- Martin H. Groschup
- Stephan Günther
- Alexandro Guterres
- Roy A. Hall
- John Hammond
- Mohamed Hassan
- Jussi Hepojoki
- Satu Hepojoki
- Udo Hetzel
- Roger Hewson
- Bernd Hoffmann
- Seiji Hongo
- Dirk Höper
- Masayuki Horie
- Holly R. Hughes
- Timothy H. Hyndman
- Amara Jambai
- Rodrigo Jardim
- Dàohóng Jiāng
- Qi Jin
- Gilda B. Jonson
- Sandra Junglen
- Serpil Karadağ
- Karen E. Keller
- Boris Klempa
- Jonas Klingström
- Gary Kobinger
- Hideki Kondō
- Eugene V. Koonin
- Mart Krupovic
- Gael Kurath
- Ivan V. Kuzmin
- Lies Laenen
- Robert A. Lamb
- Amy J. Lambert
- Stanley L. Langevin
- Benhur Lee
- Elba R. S. Lemos
- Eric M. Leroy
- Dexin Li
- Jiànróng Lǐ
- Mifang Liang
- Wénwén Liú
- Yàn Liú
- Igor S. Lukashevich
- Piet Maes
- William Marciel de Souza
- Marco Marklewitz
- Sergio H. Marshall
- Giovanni P. Martelli
- Robert R. Martin
- Shin-Yi L. Marzano
- Sébastien Massart
- John W. McCauley
- Nicole Mielke-Ehret
- Angelantonio Minafra
- Maria Minutolo
- Ali Mirazimi
- Hans-Peter Mühlbach
- Elke Mühlberger
- Rayapati Naidu
- Tomohide Natsuaki
- Beatriz Navarro
- José A. Navarro
- Sergey V. Netesov
- Gabriele Neumann
- Norbert Nowotny
- Márcio R. T. Nunes
- Are Nylund
- Arnfinn L. Økland
- Renata C. Oliveira
- Gustavo Palacios
- Vicente Pallas
- Bernadett Pályi
- Anna Papa
- Colin R. Parrish
- Alex Pauvolid-Corrêa
- Janusz T. Pawęska
- Susan Payne
- Daniel R. Pérez
- Florian Pfaff
- Sheli R. Radoshitzky
- Aziz-ul Rahman
- Pedro L. Ramos-González
- Renato O. Resende
- Carina A. Reyes
- Bertus K. Rima
- Víctor Romanowski
- Gabriel Robles Luna
- Paul Rota
- Dennis Rubbenstroth
- Jonathan A. Runstadler
- Daniel Ruzek
- Sead Sabanadzovic
- Jiří Salát
- Amadou Alpha Sall
- Maria S. Salvato
- Kamil Sarpkaya
- Takahide Sasaya
- Martin Schwemmle
- Muhammad Z. Shabbir
- Xiǎohóng Shí
- Zhènglì Shí
- Yukio Shirako
- Peter Simmonds
- Jana Širmarová
- Manuela Sironi
- Sophie Smither
- Teemu Smura
- Jin-Won Song
- Kirsten M. Spann
- Jessica R. Spengler
- Mark D. Stenglein
- David M. Stone
- Petra Straková
- Ayato Takada
- Robert B. Tesh
- Natalie J. Thornburg
- Keizō Tomonaga
- Noël Tordo
- Jonathan S. Towner
- Massimo Turina
- Ioannis Tzanetakis
- Rainer G. Ulrich
- Anna Maria Vaira
- Bernadette van den Hoogen
- Arvind Varsani
- Nikos Vasilakis
- Martin Verbeek
- Victoria Wahl
- Peter J. Walker
- Hui Wang
- Jianwei Wang
- Xifeng Wang
- Lin-Fa Wang
- Tàiyún Wèi
- Heather Wells
- Anna E. Whitfield
- John V. Williams
- Yuri I. Wolf
- Zhìqiáng Wú
- Xin Yang
- Xīnglóu Yáng
- Xuejie Yu
- Natalya Yutin
- F. Murilo Zerbini
- Tong Zhang
- Yong-Zhen Zhang
- Guohui Zhou
- Xueping Zhou
Corresponding author
Correspondence toJens H. Kuhn.
Ethics declarations
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of the Army, the US Department of Defense, the US Department of Health and Human Services, the US Department of Homeland Security (DHS) Science and Technology Directorate (S&T), or of the institutions and companies affiliated with the authors. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. In no event shall any of these entities have any responsibility or liability for any use, misuse, inability to use, or reliance upon the information contained herein. The US departments do not endorse any products or commercial services mentioned in this publication. The US Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.
Funding
This work was supported in part through Laulima Government Solutions, LLC prime contract with the US National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272201800013C. J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC under Contract No. HHSN272201800013C. This project has been funded in whole or in part with federal funds from the National Cancer Institute (NCI), National Institutes of Health (NIH), under Contract No. 75N91019D00024, Task Order No. 75N91019F00130 to I.C., who was supported by the Clinical Monitoring Research Program Directorate, Frederick National Lab for Cancer Research, sponsored by NCI. This work was also funded in part by Contract No. HSHQDC-15-C-00064 awarded by the US Department of Homeland Security (DHS) Science and Technology Directorate (S&T) for the management and operation of The National Biodefense Analysis and Countermeasures Center (NBACC), a federally funded research and development center operated by the Battelle National Biodefense Institute (V.W.); and NIH contract HHSN272201000040I/HHSN27200004/D04 and grant R24AI120942 (N.V., R.B.T.). S.S. acknowledges partial support from the Special Research Initiative of Mississippi Agricultural and Forestry Experiment Station (MAFES), Mississippi State University, and the National Institute of Food and Agriculture, US Department of Agriculture, Hatch Project 1021494.
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Tim Skern and Sead Sabanadzovic.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kuhn, J.H., Adkins, S., Alioto, D. et al. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales.Arch Virol 165, 3023–3072 (2020). https://doi.org/10.1007/s00705-020-04731-2
- Received: 15 June 2020
- Accepted: 04 July 2020
- Published: 04 September 2020
- Issue Date: December 2020
- DOI: https://doi.org/10.1007/s00705-020-04731-2