Altered Gut Flora Are Associated with Septic Complications and Death in Critically Ill Patients with Systemic Inflammatory Response Syndrome (original) (raw)
Abstract
Background
Gut under severe insult is considered to have an important role in promoting infection and multiple organ dysfunction syndrome from the viewpoint of altered intestinal epithelium, immune system and commensal bacteria. There are few reports, however, about the relationship between gut flora and septic complications.
Methods
We analyzed gut flora in patients with systemic inflammatory response syndrome (SIRS) and evaluated key bacteria and their cutoff values for infectious complications and mortality by using classification and regression trees (CART). Eighty-one SIRS patients with a serum C-reactive protein level higher than 10 mg/dL treated in the intensive care unit (ICU) for more than 2 days were included for the study. We quantitatively evaluated nine types of bacteria in fecal samples by plate or tube technique. Two hundred seventy-one samples were analyzed using CART and logistic regression.
Results
The dominant factors for complication of enteritis were the minimum number of total obligate anaerobes and the maximum number of Staphylococcus and Enterococcus. The dominant factors for complication of bacteremia were the minimum numbers of total obligate anaerobes and total facultative anaerobes. The dominant factors for mortality were the numbers of total obligate anaerobes and total facultative anaerobes and age.
Conclusions
A decrease in total obligate anaerobes and an increase in pathogenic bacteria in the gut are associated with septic complications and mortality in patients with SIRS. The altered gut flora may be a potential prognostic marker in SIRS patients.
Similar content being viewed by others
Introduction
The cross talk between host and gut flora has a major influence on host physiology in health and disease. Commensal gut flora have important and specific metabolic, trophic and protective functions. Equilibrium between species of resident bacteria provides stability of the microbial population within an individual under normal conditions [1]. Maintaining the balance of gut flora is important for a functional immune system. Recent studies have shown that human intestinal microflora contain at least 100 times as many genes as the human genome [2] and that humans are superorganisms for which metabolism involves an amalgamation of microbial and human activities. Macpherson et al. [3] reported that commensal bacteria are carried to the mesenteric lymph nodes by intestinal dendritic cells. Dendritic cells induce IgA production, which helps protect against mucosal penetration by commensal bacteria. Rakoff-Nahoum et al. [4] reported that recognition of commensal microflora by Toll-like receptors is required for increased proliferation of colonic epithelial cells and protection against dextran sulfate sodium induced intestinal epithelial injury. These reports suggest that the commensal gut flora in humans play an essential role in homeostasis and protection from injury in the gut.
Under normal circumstances in the gut there are approximately one hundred billion colony forming unit (CFU) bacteria in 1 g of feces, and most are total obligate anaerobes such as Bacteroidaceae and Bifidobacterium. Hooper et al. [5] colonized germ-free mice with Bacteroides thetaiotaomicron and compared gene expression in germ-free and colonized mice by DNA microarray analysis. Results showed that the increased expressions of genes were involved in nutrient absorption, mucosal barrier fortification, xenobiotic metabolism, angiogenesis and postnatal intestinal maturation. Breast-fed infants seem to develop a flora rich in Bifidobacterium in comparison to formula-fed babies and are less often colonized with bacteria such as Staphylococcus and Escherichia coli [6]. These findings indicate that indigenous obligate anaerobes seem to play an essential role from the beginning in establishing the balance of the gut flora and other physiological functions.
It has been reviewed that various diseases are associated with a change in gut flora [7]. Studies of diseases and abnormal gut flora have focused on various diseases, such as inflammatory bowel disease [8], atopic asthma [9], obesity [10], and hepatic encephalopathy [11]. Some diseases are associated with a decrease in Bifidobacterium [12]. We recently evaluated microflora (bacterial counts of 10 key groups) and environmental changes (pH and nine organic acids) in patients with severe systemic inflammatory response syndrome (SIRS) and showed significant alteration in patients with severe SIRS [13]. Patients with SIRS showed 100–10,000 fewer total obligate anaerobes and 2–4 log units fewer beneficial Bifidobacterium than healthy volunteers. The disruption of commensal gut flora may affect gut barrier function and the systemic inflammatory response following severe insult.
In this study, we analyzed gut flora in 81 patients with severe SIRS and examined whether the gut flora are associated with infectious complications and prognosis by using classification and regression trees (CART).
Methods
Patients
Subjects included 81 patients with severe SIRS who were admitted to the Department of Traumatology and Acute Critical Medicine, Osaka University Medical School, Osaka, Japan and treated in the intensive care unit (ICU) for more than 2 days during the period of December 2003 to April 2006. Severe SIRS was diagnosed according to the criteria of the American College of Chest Physicians and the Society of Critical Care Medicine [14] and in patients with a serum C-reactive protein level greater than 10 mg/dL. Enteral nutrition (Impact, Novartis Nutrition, Minneapolis, MN) was initiated as soon as possible. If infections occurred, patients were initially treated empirically for the underlying clinical syndrome and then according to the results of antibiotics susceptibility testing of the isolated bacterial infection. Antibiotics were administered with the same policy for all study periods. A histamine H2 receptor blocker (20 mg famotidine) was administered routinely twice daily until the initiation of oral intake. After admission, fecal samples were collected serially and analyzed. This study was approved by the Institutional Review Board of Osaka University, and informed consent was obtained from the family of each patient.
Fecal Bacteriologic Culture
Feces were collected in a test tube, which was maintained under anaerobic conditions in an atmosphere of 7% H2 and 5% CO2 in N2. The test tube was cooled in an icebox before culture. VL-G roll tube agar [15] supplemented with 0.2% cellobiose and 0.2% maltose (modified VL-G roll tube agar) was used to determine total anaerobe counts. Different media were used for selective isolation of different microorganisms: modified VL-G roll tube agar to which 80 μg/mL vancomycin and 1 μg/mL kanamycin were added for Bacteroidaceae; CW agar (Nikken Bio Medical Laboratory Inc., Kyoto, Japan) for lecithinase-positive Clostridium; MPN agar [16] for Bifidobacterium; COBA agar for Enterococcus; LBS agar (Becton Dickinson and Company, Cockeysville, MD) supplemented with 0.8% Laboratory Lemco powder (Oxoid Co. Ltd., Basingtoke, UK) for Lactobacillus; Staphylococcus medium no. 110 agar (Nissui Pharmaceutical Co. Ltd., Tokyo, Japan) for Staphylococcus; DHL agar (Nissui Pharmaceutical Co. Ltd.) for Enterobacteriaceae; NAC agar (Nissui Pharmaceutical Co. Ltd.) for Pseudomonas; and GS agar (Nissui Pharmaceutical Co. Ltd.) for Candida [17, 18]. CW agar and LBS agar were cultured anaerobically at 37°C for 72 h. After incubation, colonies on plates were counted and gram stained. Numbers of viable bacteria per gram of feces (wet weight) were calculated. All bacterial counts (colony-forming units (CFU)/g of wet feces) were transformed to logarithm (log10CFU) for ease of statistical analysis. The lower limit of bacterial detection with this procedure was 1,000 CFU per gram of feces for the obligate anaerobes, Bacteroidaceae and Bifidobacterium, and 100 CFU per gram of feces for other bacteria. The reproducibility and stability of these measurements have previously been shown.
Surveillance and Definition of Infection
Body temperature was measured continuously. Surveillance cultures from urine, blood, and sputum were performed routinely for each patient. In cases of suspected infection, laboratory testing, chest X-ray, and computed tomography scanning were performed as necessary. Enteritis was diagnosed in accordance with the definition of the National Nosocomial Infections Surveillance system [[19](/article/10.1007/s10620-010-1418-8#ref-CR19 "Centers for Disease Control and Prevention: National Nosocomial Infections Surveillance System (NNIS). Available at: http://www.cdc.gov/ncidod/dhqp/nnis_pubs.html
. Accessed December 12, 2009.")\]. Bacteremia was defined as a positive blood culture. These new infectious complications and mortality were evaluated within 60 days from admission.Statistical Analysis
Analysis was performed using infectious complications and mortality as the binary outcome variable. Age, sex, APACHE II score on admission, the usage of antibiotics and nine species of bacteria in the fecal samples were used as potential predictor covariates. Classification and regression trees (CART) is a binary recursive partitioning using nonparametric approaches. It is a popular method for identifying high-risk and poor prognosis groups. The overall study group is split into two subgroups using the most powerful predictor of the outcome. This splitting is repeated within the subgroups until no further significant splits are found or the subgroups become too small. The results are displayed in the binary trees structure which, in a final step, is pruned as necessary. CART was used in order to determine a key bacterium and the cutoff value for infectious complications and mortality [20]. Ordinary and exact logistic regressions were performed to evaluate quantitatively the effect of the covariates that were suggested by analysis of CART.
To evaluate the changes of fecal flora, we represented these changes for the minimum, maximum and average of each patient. The method is as follows. First, the minimum, maximum, and average data of each patient were selected from all fecal bacterial data. In the case of mortality, the last data were selected for independent covariates additionally. Second, recursive partitioning was used to create a tree using the whole independent covariates. Third, we selected three predominant covariates and made a logistic regression model. Predictive accuracy of CART was evaluated using sensitivity, specificity for infectious complications (enteritis and bacteremia) and mortality. All statistical tests were two-sided at a significance level of 0.05. Statistical analysis was performed with CART version 6.0 (Salford Systems), StatXact version 8.0 (Cytel Inc.) and SAS version 9.1 (SAS Institute Inc.).
Results
Patient characteristics are listed in Table 1. The study group was comprised of 48 men and 33 women with a mean (±SD) age of 57.4 ± 20.3 years. The origin of severe SIRS was sepsis in 50 patients, trauma in 23 patients, and major burns in eight patients. Acute Physiology and Chronic Health Evaluation (APACHE II score) on admission was 17.1 ± 7.1. The number of antibiotics type was 4.3 ± 2.6, and the duration of antibiotics usage was 19.6 ± 20.0 days. Infectious complications such as enteritis and bacteremia occurred in 22 and 23 patients, respectively. Mortality was 30.9%. Analysis of blood cultures showed Staphylococcus species in 15 patients, P. aeruginosa in six patients, Enterococcus faecalis in four patients, Escherichia coli in three patients and others.
Table 1 Patients characteristics with severe systemic inflammatory response syndrome (SIRS) (n = 81)
The total number of fecal samples was 271. Each patient had 3.3 ± 2.8 samples, and the date when the sample was taken was 21.1 ± 15.6 days after admission (mean ± SD). Characteristics of the minimum, maximum, and average data from each patient are shown in Table 2. CART and multivariate logistic regression of the predominant covariates were developed for the following dependent covariates: enteritis, bacteremia, and mortality (Table 3).
Table 2 Characteristics of gut flora (n = 81)
Table 3 Results of multivariate logistic regression analysis
Enteritis
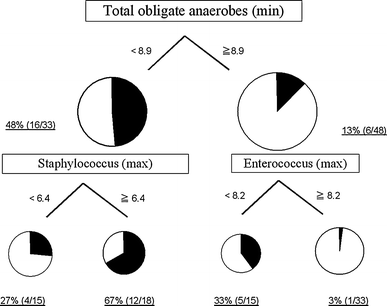
The primary split was determined to be the minimum number of total obligate anaerobes and the cutoff value was 8.9 (log10CFU/g). Further partitioning was based on the maximum number of Staphylococcus and Enterococcus. The incidences of enteritis in each partition are shown in Fig. 1. The diagnostic characteristics of this tree are sensitivity 77.3% and specificity 72.9%.
Fig. 1
Incidences of enteritis partitioned by total obligate anaerobes (min), Staphylococcus (max), and Enterococcus (max) using CART. Total obligate anaerobes (min), the minimum number of total obligate anaerobes; Staphylococcus (max), the maximum number of Staphylococcus; Enterococcus (max), the maximum number of Enterococcus; CART, classification and regression trees
Bacteremia
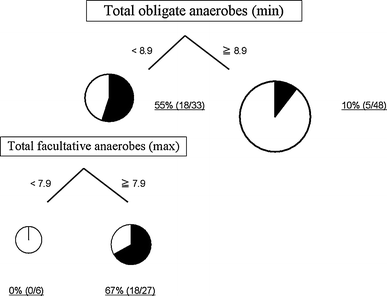
The primary split was determined to be the minimum number of total obligate anaerobes and the cutoff value was 8.9 (log10CFU/g). Further partitioning was based on the maximum number of total facultative anaerobes. The incidences of bacteremia in each partition are shown in Fig. 2. The diagnostic characteristics of this tree are sensitivity 69.6% and specificity 84.5%.
Fig. 2
Incidences of bacteremia partitioned by total obligate anaerobes (min) and total facultative anaerobes (min) using CART. Total obligate anaerobes (min), the minimum number of total obligate anaerobes; Total facultative anaerobes (max), the maximum number of total facultative anaerobes; CART, classification and regression trees
Mortality
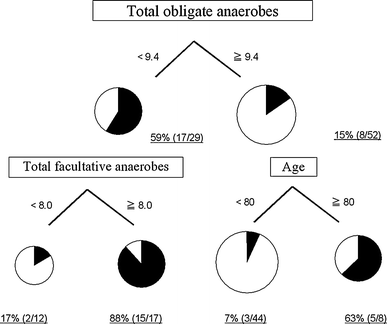
The primary split was determined to be the number of total obligate anaerobes at the last sample and the cutoff value was 9.4 (log10CFU/g). Further partitioning was based on total facultative anaerobes and age. The mortality in each partition is shown in Fig. 3. The diagnostic characteristics of this tree are sensitivity 80.0% and specificity 91.1%.
Fig. 3
Mortality partitioned by total obligate anaerobes, total facultative anaerobes, and age using CART. Total obligate anaerobes, the number of total obligate anaerobes; Total facultative anaerobes, the number of total facultative anaerobes at the last sample; CART, classification and regression trees
The APACHE II score on admission was 18.2 ± 7.1 in patients with total obligate anaerobes ≤9.4 (log10CFU/g), and 18.1 ± 13.4 in patients with total obligate anaerobes ≧9.4 (log10CFU/g) (mean ± SD). There was no significant difference in APACHE II score between the two groups. In patients with total obligate anaerobes ≤9.4 (log10CFU/g), the APACHE II score was 17.1 ± 5.2 in patients with total facultative anaerobes ≤8.0 (log10CFU/g), and 18.9 ± 8.2 in patients with total facultative anaerobes ≧8.0 (log10CFU/g) (mean ± SD). There was no significant difference in APACHE II score between the two groups. In patients with total obligate anaerobes ≧9.4 (log10CFU/g), the APACHE II score was 18.6 ± 14.6 in patients ≤80 years of age, and 15.5 ± 6.3 in patients ≧80 years of age (mean ± SD). There was no significant difference in APACHE II score between the two groups. The APACHE II score on admission was not selected as a major covariate by CART analysis.
Discussion
This study provides the first in vivo evidence that an altered gut flora is associated with infectious complications and mortality in patients with severe SIRS using CART and logistic regression.
CART is a commonly used method for identifying high-risk and poor prognosis groups. We selected this method because we wanted to identify both the key bacteria for infectious complications and mortality, and to determine the utility of the partition number as a clinical marker. In this study, the most predictive factor for enteritis and bacteremia was the minimum number of total obligate bacteria. The number of obligate anaerobes in the last sample was the most predictive factor for mortality. These results showed that alteration of the gut flora, not only the facultative anaerobes but also the total obligate anaerobes, is a potential prognostic factor in patients with severe SIRS.
Obligate anaerobes inhibit the growth of other bacteria, a phenomenon referred to as “colonization resistance” [21]. Previous animal studies have shown a very low incidence of bacterial translocation when obligate anaerobes are maintained in the gut, suggesting that obligate anaerobic bacteria are the principal inhibitors of bacterial overgrowth and translocation of potentially pathogenic bacteria [22].
In this study, the minimum number of total facultative anaerobes was also one of the dominant covariates for bacteremia. In our previous study, analysis of fecal flora showed that SIRS patients had markedly lower total anaerobic bacterial counts (particularly of beneficial Bifidobacterium and Lactobacillus) and higher total facultative anaerobes such as Staphylococcus and Pseudomonas counts than those of healthy volunteers [13]. In patients who showed bacterial translocation, Gram-negative facultative anaerobes were the most common organisms identified in mesenteric lymph nodes and serosal scrapings at laparotomy; obligate anaerobes were cultured less frequently [23]. These findings indicate that the equilibrium between obligate anaerobes and total facultative anaerobes plays a critical role in causing septic complications in severely ill patients. A decrease in obligate anaerobes and an increase in total facultative anaerobes may weaken intestinal resistance to pathogens. Since antibiotics were used in most patients in our study, further study is needed to evaluate if the altered gut flora was mainly due to severe SIRS or other factors such as antibiotics.
The age was detected as one of the dominant covariates for mortality even when the number of total obligate anaerobes was within the normal range. Previous reports have shown that immune functions decrease with ageing [24]. The composition of the gut flora shifts with aging, with increased facultative anaerobes and decreased beneficial bacteria such as the Lactobacillus and Bifidobacterium, which have been reported in studies of elderly people compared with younger people [25], implying a more fragile balance of the gut flora. There is evidence showing that, when ageing, the barrier function of the gut weakens with increased gut permeability [26]. These findings indicate that the combination of the weakened overall immune defense together with the fragile intestinal flora balance could have resulted in the finding that the elderly could easily develop enteritis under critically ill conditions even when the number of total obligate anaerobes was normal.
As for enteritis, the first split was the number of the minimum of total obligate anaerobes, which was the same as found for bacteremia. The second split was the maximum number of Staphylococcus. The increase in S. aureus and the decrease in total obligate anaerobes was an important predictive factor for the progression of bacteremia. The third split was the number of the maximum of Enterococcus. Enterococcus is generally considered as having an ambiguous status concerning its safety assessment procedure. On the one hand, Enterococcus is a natural member of the digestive microbiota in humans and can produce bacteriocins against other virulent bacteria and has a protective influence on the gut. On the other hand, Enterococcus is considered as an emerging pathogen for humans [27]. Further clarification is required about the impact of the number of Enterococcus on enteritis.
This study did not aim at making an exact formula for septic complications, but at finding out bacteria of high risk. The alteration of normal gut flora may be more important for development of septic complications than the increase in pathogenic bacteria. The next target for our study is to maintain gut flora and in this way attain any associated clinical benefits. Further study is needed to clarify the mechanism by which a decrease in total obligate anaerobes induces lethal infectious complications and mortality in SIRS patients.
In conclusion, a decrease in total obligate anaerobes and an increase in pathogenic bacteria in the gut are associated with septic complications and high mortality in patients with severe SIRS, and these would be potential prognostic markers of patient survival.
References
- Guarner F, Malagelada J. Gut flora in health and disease. Lancet. 2003;361:512–519.
Article PubMed Google Scholar - Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359.
Article PubMed CAS Google Scholar - Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665.
Article PubMed CAS Google Scholar - Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241.
Article PubMed CAS Google Scholar - Hooper LV, Wong MH, Thelin A, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884.
Article PubMed CAS Google Scholar - Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–67.
Article PubMed CAS Google Scholar - Guarner F. Enteric flora in health and disease. Digestion. 2006;73(Suppl 1):5–12.
Article PubMed Google Scholar - Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594.
Article PubMed CAS Google Scholar - Bjorksten B. Primary prevention of atopic asthma. Curr Opin Allergy Clin Immunol. 2001;1:545–548.
Article PubMed CAS Google Scholar - Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023.
Article PubMed CAS Google Scholar - Liu Q, Duan Z, Ha da K, et al. Synbiotic modulation of gut flora: effect on minimal hepatic encephalopathy in patients with cirrhosis. Hepatology. 2004;39:1441–1449.
Article PubMed Google Scholar - Kerckhoffs AP, Samsom M, van der Rest ME, et al. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol. 2009;15:2887–2892.
Article PubMed Google Scholar - Shimizu K, Ogura H, Goto M, et al. Altered gut flora and environment in patients with severe SIRS. J Trauma. 2006;60:126–133.
Article PubMed Google Scholar - American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med. 1992;20:864-874.
Google Scholar - Azuma R, Suto T. Validity of transfer of the taxonomical position of Corynebacterium pseudopyogenes from genus Corynebacterium to genus Actinomyces. In: Izuka H, Hasegawa T, eds. Proceeding of the First International Conference on Culture Collection. Tokyo: University of Tokyo Press; 1970:493–505.
Google Scholar - Tanaka R, Mutai M. Improved medium for selective isolation and enumeration of Bifidobacterium. Appl Environ Microbiol. 1980;40:866–869.
PubMed CAS Google Scholar - Petts DN. Colistin-oxolinic acid-blood agar: a new selective medium for streptococci. J Clin Microbiol. 1984;19:4–7.
PubMed CAS Google Scholar - Mitsuoka T, Ono K. Fecal flora of man V communication: the fluctuation of the fecal flora of the healthy adult (author’s transl). Zentralbl Bakteriol [OrigA]. 1977;238:228–236.
CAS Google Scholar - Centers for Disease Control and Prevention: National Nosocomial Infections Surveillance System (NNIS). Available at: http://www.cdc.gov/ncidod/dhqp/nnis_pubs.html. Accessed December 12, 2009.
- Breiman L, Friedman JF, Olshen RA, et al. Classification and Regression Trees. New York: Chapman & Hall; 1984.
Google Scholar - Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409–414.
PubMed CAS Google Scholar - Steffen EK, Berg RD, Deitch EA. Comparison of translocation rates of various indigenous bacteria from the gastrointestinal tract to the mesenteric lymph node. J Infect Dis. 1988;157:1032–1038.
PubMed CAS Google Scholar - O’Boyle CJ, MacFie J, Mitchell CJ, et al. Microbiology of bacterial translocation in humans. Gut. 1998;42:29–35.
Article PubMed Google Scholar - Danielle AW, Silva AB, Palmer DB. Immunosenescence: emerging challenges for an ageing population. Immunology. 2007;120:435–446.
Article Google Scholar - Woodmansey EJ. Intestinal bacteria and ageing. J Appl Microbiol. 2007;102:1178–1186.
Article PubMed CAS Google Scholar - Saltzman JR, Kowdley KV, Perrone G, et al. Changes in small-intestine permeability with aging. J Am Geriatr Soc. 1995;43:160–164.
PubMed CAS Google Scholar - Ogier JC, Serror P. Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol. 2008;126:291–301.
Article PubMed CAS Google Scholar
Disclosure of financial arrangements
This work was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
- Department of Clinical Quality Management, Osaka University Hospital, Osaka, Japan
Kentaro Shimizu - Department of Traumatology and Acute Critical Medicine, Osaka University Graduate School of Medicine, 2-15 Yamadaoka, Suita-city, Osaka, 565-0871, Japan
Kentaro Shimizu, Hiroshi Ogura, Miki Goto, Osamu Tasaki & Yasuyuki Kuwagata - Department of Biomedical Statistics, Osaka University Graduate School of Medicine, Osaka, Japan
Toshimitsu Hamasaki - Yakult Central Institute for Microbiological Research, Tokyo, Japan
Takashi Asahara, Koji Nomoto & Masami Morotomi - Department of Trauma and Critical Care Medicine, Social Insurance Chukyo Hospital, Nagoya, Aichi, Japan
Asako Matsushima - Hoshigaoka Koseinenkin Hospital, Hirakata, Osaka, Japan
Hisashi Sugimoto
Authors
- Kentaro Shimizu
You can also search for this author inPubMed Google Scholar - Hiroshi Ogura
You can also search for this author inPubMed Google Scholar - Toshimitsu Hamasaki
You can also search for this author inPubMed Google Scholar - Miki Goto
You can also search for this author inPubMed Google Scholar - Osamu Tasaki
You can also search for this author inPubMed Google Scholar - Takashi Asahara
You can also search for this author inPubMed Google Scholar - Koji Nomoto
You can also search for this author inPubMed Google Scholar - Masami Morotomi
You can also search for this author inPubMed Google Scholar - Asako Matsushima
You can also search for this author inPubMed Google Scholar - Yasuyuki Kuwagata
You can also search for this author inPubMed Google Scholar - Hisashi Sugimoto
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toKentaro Shimizu.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Shimizu, K., Ogura, H., Hamasaki, T. et al. Altered Gut Flora Are Associated with Septic Complications and Death in Critically Ill Patients with Systemic Inflammatory Response Syndrome.Dig Dis Sci 56, 1171–1177 (2011). https://doi.org/10.1007/s10620-010-1418-8
- Received: 12 December 2009
- Accepted: 31 August 2010
- Published: 08 October 2010
- Issue Date: April 2011
- DOI: https://doi.org/10.1007/s10620-010-1418-8