International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity (original) (raw)
Abstract
Beginning in 1970, a committee was constituted under the auspices of the World Health Organization (WHO) to catalog primary immunodeficiencies. Twenty years later, the International Union of Immunological Societies (IUIS) took the remit of this committee. The current report details the categorization and listing of 354 (as of February 2017) inborn errors of immunity. The growth and increasing complexity of the field have been impressive, encompassing an increasing variety of conditions, and the classification described here will serve as a critical reference for immunologists and researchers worldwide.
Similar content being viewed by others
Introduction
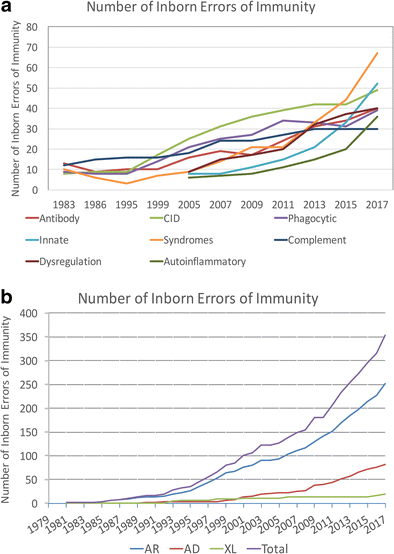
In 1970, Drs. Fudenberg, Good, Hitzig, Kunkel, Roitt, Rosen, Rowe, Seligmann, and Soothill met under the auspices of the World Health Organization to classify the emerging “primary immune deficiencies.” This august group focused on understanding whether immunodeficiencies could be categorized as B cell disorders or T cell disorders [1, 2]. Their initial report identified 16 distinct immunodeficiencies and included the prophetic comment that “the variable immunodeficiency group probably lumps together a series of syndromes…. Included in this group are cases previously classified as ‘congenital’, non-sex linked or sporadic hypogammaglobulinemia, primary ‘dysgammglobulinemia’ of both childhood and adult life, and ‘acquired’ primary hypogammaglobulinemia. It is hoped that careful analysis of such patients…. will result in delineation of several homogeneous syndromes…”. Indeed, the emergence of monogenic causes of hypogammaglobulinemia (Table 3) and disorders with variable immunoglobulin abnormalities associated with immune dysregulation (Table 4) have been the groups of immunodeficiencies most transformed by the advent of new technologies. Another group dramatically impacted by resetting of the clinical radar and new techniques has been the set of disorders associated with a limited spectrum of infectious susceptibility. The graphs in Fig. 1 define the transformation of the field over the interval during which next-generation sequencing came to prominence. The tremendous progress, energy, and enthusiasm in the field currently have led to a greater need than ever for a current cataloging of the disorders.
Fig. 1
Each publication of the World Health Organization and IUIS Primary Immunodeficiencies Committee was reviewed for the number of conditions listed and displayed graphically [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19]. The rapid increase in the twenty-first century relates to improved awareness and increasing use of sequencing. Assuming 20,000 coding genes in the human genome, inborn errors of immunity are implicated through mutations in 1.7% of these genes. There are now 330 specific disorders, 320 monogenic defects, 312 distinct genes (nine genes with both LOF and GOF and C4 deficiency requiring defects in both C4A and C4B). a The categorization of the inborn errors of immunity according the schema in the current manuscript. b The categorization of the inborn errors of immunity according to their inheritance
The new disorders (since 2015 [3]) represent an impressive spectrum of phenotypes. There are 354 distinct disorders with 344 different gene defects listed. The emerging dominance of next-generation sequencing has driven the rapid increase in the number of recognized disorders which has led to two major consequences. Often new inborn errors of immunity are initially described in a single kindred or a small number of kindreds. This may lead to incorrect assumptions about prevalence and phenotype. In fact, for most disorders, we have little idea of the prevalence within even the recognized population with the described phenotype. The second consequence of the rapid rise of next-generation sequencing is a striking expansion of the phenotypic spectrum associated with many diseases. Where once the phenotype of a given disorder was clear, the spectrum of manifestations often extends impressively once the ascertainment is not linked to a preconceived idea [20]. As a community, we recognize the importance of publishing cases and small series and to report specific mutations with clinical findings because publications are used to define likelihood of causality during bioinformatic analysis of next-generation sequencing results.
In 1999, the Committee on Primary Immunodeficiencies came under the auspices of the International Union of Immunological Societies (IUIS). The current committee met on February 23–24, 2017, in London to update the classification of human primary immunodeficiencies. Inclusion in this “master list” requires a body of literature supporting causality of a gene defect and a penetrance indicating clinical relevance [21]. Committee members vote on inclusion of each new disorder and this publications lists those included as of the February 2017 meeting. The landscape is changing so rapidly, and the number of primary immunodeficiencies growing so fast, that two major changes have been implemented. The published list will continue to serve as a reference; however, this list will now be available as a csv file on the IUIS website to enable sorting according to gene, disease name, or clinical/laboratory feature. This file will also include the associated ICD10 codes in order to promote harmonization of utilization. The second major change is to the nomenclature. The term primary immunodeficiency has an important legacy—the abbreviations PID or PIDD are often used by patient organizations and are recognized around the world. However, this terminology does limit the conceptualization of disorders to those in which susceptibility to infection is the main manifestation. The improving recognition of immune dysregulation diseases, including the growing field of autoinflammatory disorders and interferonopathies, has mandated that a more encompassing terminology be used. This manuscript, therefore, utilizes “inborn errors of immunity” as the descriptor for the work and the categorization. In addition to embracing technology to remain updated, the companion publication “Update of the Phenotypical IUIS Classification for Primary Immunodeficiencies” will provide a phenotype-oriented approach to the IUIS categorization of disorders. Moreover, a new free application can be found as “PID phenotypical diagnosis” or “PID classification” from iTunes and Android app stores [[22](/article/10.1007/s10875-017-0464-9#ref-CR22 "Boufiha A, Phenotypical classification of PIDD. 2017. https://play.google.com/store/apps/details?id=com.horiyasoft.pidclassification
."), [23](/article/10.1007/s10875-017-0464-9#ref-CR23 "Bousfiha A, Phenotypical classification of PIDD, iTunes. 2017.
https://itunes.apple.com/us/app/pid-phenotypical-diagnosis/id1160729399?mt=8
.")\]. Information that is readily accessible is the new standard, and the IUIS Expert Committee on Primary Immunodeficiencies believes that improved access to information will positively impact patient care around the world.The tables divide disease categories according to common phenotypes for ease of review and searching. Table 1 lists combined immunodeficiencies, Table 2 lists combined immunodeficiencies with syndromic features, Table 3 lists predominantly antibody deficiencies, Table 4 lists diseases of immune dysregulation, Table 5 lists defects of phagocyte number or function, Table 6 lists defects in intrinsic and innate immunity, Table 7 lists autoinflammatory diseases, Table 8 lists complement deficiencies, and Table 9 lists phenocopies of inborn errors of immunity. The division into phenotypes for the purpose of this list does not imply that the presentation is homogeneous. Each disorder is listed only once for the sake of simplicity although distinct modes of inheritance can be listed separately. There are nine genes for which both loss-of-function and gain-of-function variants have been identified: CFB, C3, CARD11, STAT1, STAT3, WAS, JAK1, IFIH1, and ZAP70. For these, the loss-of-function and gain-of-function aspects are listed. Within each table, there are additional sub-tables that segregate into coherent phenotypic sets. At the end of each table, the new disorders, added for this publication, are listed for easy reference. Other features important for navigation of the list include the use of OMIM links [[24](/article/10.1007/s10875-017-0464-9#ref-CR24 "OMIM Online Mendelian Inheritance in Man. https://www.ncbi.nlm.nih.gov/omim
.")\]. For additional information on a gene, the links can be accessed from within the online publication. For the second time, we also include non-inborn errors of immunity in Table [9](/article/10.1007/s10875-017-0464-9#Tab9), representing phenocopies of inborn errors which might be important to consider diagnostically.Table 1 Immunodeficiencies affecting cellular and humoral immunity
Table 2 Combined immunodeficiencies with associated or syndromic features
Table 3 Predominantly antibody deficiencies
Table 4 Diseases of immune dysregulation
Table 5 Congenital defects of phagocyte number or function
Table 6 Defects in intrinsic and innate immunity
Table 7 Autoinflammatory disorders
Table 8 Complement deficiencies
Table 9 Phenocopies of inborn errors of immunity
The goal of the IUIS Expert Committee on Primary Immunodeficiencies is to increase awareness, facilitate recognition, promote optimal treatment, and support research in the field of immune deficiency disorders. Thus, the “IUIS PID Committee Report on Inborn Errors of Immunity” and “Update of the Phenotypical IUIS Classification for Primary Immunodeficiencies” publications are important resources for clinicians and researchers. In addition, these tables form the basis of lists used for sequencing panels and are used to monitor health utilization which will influence health services funding by federal or state governments and/or insurance companies in various global settings. The addition of ICD10 codes for the online version will promote a harmonization between the diagnostic tables and coding items that will facilitate bioinformatics research going forward.
References
- Fudenberg H, Good RA, Goodman HC, Hitzig W, Kunkel HG, Roitt IM, et al. Primary immunodeficiencies. Report of a World Health Organization Committee. Pediatrics. 1971;47:927–46.
CAS PubMed Google Scholar - Fudenberg HH, Good RA, Hitzig W, Kunkel HG, Roitt IM, Rosen FS, et al. Classification of the primary immune deficiencies: WHO recommendation. N Engl J Med. 1970;283:656–7.
Article CAS PubMed Google Scholar - Picard C, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, et al. Primary immunodeficiency diseases: an update on the classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J Clin Immunol. 2015;35:696–726.
Article CAS PubMed PubMed Central Google Scholar - Anonymous. Primary immunodeficiency diseases. Report of a WHO scientific group. Clin Exp Immunol. 1997;109(Suppl 1):1–28.
Google Scholar - Chandra R, Cooper M, Hitzig W, Rosen F, Seligmann M, Soothill JF, et al. WHO scientific group on immunodeficiencies. Clin Immunol Immunopathol. 1979;13:296–359.
Article Google Scholar - Gatti R. On the classification of patients with primary immunodeficiency disorders. Clin Immunol Immunopathol. 1974;3:243–7.
Article CAS PubMed Google Scholar - Cooper M, Faulk W, Fudenberg H, Good R, Hitzig W, Kunkel H, et al. Meeting report of the second international workshop on primary immunodeficiency diseases in man. Clin Immunol Immunopathol. 1974;2:416–45.
Article CAS PubMed Google Scholar - Anonymous. Primary immunodeficiency diseases. Report of an IUIS Scientific Committee. International Union of Immunological Societies. Clin Exp Immunol. 1999;118(Suppl 1):1–28.
Google Scholar - Immunodeficiency WSGo: meeting report primary immunodeficiency diseases. Clinical Immunology and Immunopathology 28: 450–475, 1983.
- Rosen F, Wedgewood R, Eibl M. Primary immunodeficiency diseases. Clin Immunol Immunopathol. 1986;40:166–96.
Article Google Scholar - Bousfiha A, Jeddane L, Al-Herz W, Ailal F, Casanova JL, Chatila T, et al. The 2015 IUIS phenotypic classification for primary immunodeficiencies. J Clin Immunol. 2015;35:727–38.
Article PubMed PubMed Central Google Scholar - Bousfiha AA, Jeddane L, Ailal F, Al Herz W, Conley ME, Cunningham-Rundles C, et al. A phenotypic approach for IUIS PID classification and diagnosis: guidelines for clinicians at the bedside. J Clin Immunol. 2013;33:1078–87.
Article CAS PubMed PubMed Central Google Scholar - Primary immunodeficiency diseases. Report of an IUIS Scientific Committee. International Union of Immunological Societies. Clin Exp Immunol 118 Suppl 1: 1–28, 1999.
- Notarangelo L, Casanova JL, Fischer A, Puck J, Rosen F, Seger R, et al. Primary immunodeficiency diseases: an update. J Allergy Clin Immunol. 2004;114:677–87.
Article CAS PubMed Google Scholar - Notarangelo L, Casanova JL, Conley ME, Chapel H, Fischer A, Puck J, et al. Primary immunodeficiency diseases: an update from the International Union of Immunological Societies Primary Immunodeficiency Diseases Classification Committee Meeting in Budapest, 2005. J Allergy Clin Immunol. 2006;117:883–96.
Article PubMed Google Scholar - Geha RS, Notarangelo LD, Casanova JL, Chapel H, Conley ME, Fischer A, et al. Primary immunodeficiency diseases: an update from the International Union of Immunological Societies Primary Immunodeficiency Diseases Classification Committee. J Allergy Clin Immunol. 2007;120:776–94.
Article PubMed PubMed Central Google Scholar - International Union of Immunological Societies Expert Committee on Primary I, Notarangelo LD, Fischer A, Geha RS, Casanova JL, Chapel H, et al. Primary immunodeficiencies: 2009 update. J Allergy Clin Immunol. 2009;124:1161–78.
Article Google Scholar - Al-Herz W, Bousfiha A, Casanova JL, Chapel H, Conley ME, Cunningham-Rundles C, et al. Primary immunodeficiency diseases: an update on the classification from the international union of immunological societies expert committee for primary immunodeficiency. Front Immunol. 2011;2:54.
Article PubMed PubMed Central Google Scholar - Primary immunodeficiency diseases. Report of a WHO Scientific Group. Clin Exp Immunol 99 Suppl 1: 1–24, 1995.
- Stray-Pedersen A, Sorte HS, Samarakoon P, Gambin T, Chinn IK, Coban Akdemir ZH, et al. Primary immunodeficiency diseases: genomic approaches delineate heterogeneous Mendelian disorders. J Allergy Clin Immunol. 2017;139:232–45.
Article PubMed Google Scholar - Casanova JL, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J Exp Med. 2014;211:2137–49.
Article CAS PubMed PubMed Central Google Scholar - Boufiha A, Phenotypical classification of PIDD. 2017. https://play.google.com/store/apps/details?id=com.horiyasoft.pidclassification.
- Bousfiha A, Phenotypical classification of PIDD, iTunes. 2017. https://itunes.apple.com/us/app/pid-phenotypical-diagnosis/id1160729399?mt=8.
- OMIM Online Mendelian Inheritance in Man. https://www.ncbi.nlm.nih.gov/omim.
Acknowledgements
The authors wish to thank Dawn Westerfer for the expert secretarial support and Ulrika Smrekova for the administrative support.
Author information
Authors and Affiliations
- Center for the Study of Immunodeficiencies, Necker Hospital for Sick Children, Assistance Publique-Hôpitaux de Paris (APHP), Paris, France
Capucine Picard - Laboratory of Lymphocyte Activation and Susceptibility to EBV, INSERM UMR1163, Imagine Institute, Necker Hospital for Sick Children, Paris Descartes University, Paris, France
Capucine Picard - UCL Great Ormond Street Institute of Child Health, London, UK
H. Bobby Gaspar - Department of Pediatrics, Faculty of Medicine, Kuwait University, Kuwait City, Kuwait
Waleed Al-Herz - Laboratoire d’Immunologie Clinique, d’Inflammation et d’Allergy LICIA Clinical Immunology Unit, Casablanca Children’s Hospital, Ibn Rochd Medical School, King Hassan II University, Casablanca, Morocco
Aziz Bousfiha - St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY, USA
Jean-Laurent Casanova - Howard Hughes Medical Institute, New York, NY, USA
Jean-Laurent Casanova - Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM UMR1163, Imagine Institute, Necker Hospital for Sick Children, University Paris Descartes, Paris, France
Jean-Laurent Casanova - Pediatric Hematology-Immunology Unit, Necker Hospital for Sick Children APHP, Paris, France
Jean-Laurent Casanova - Division of Immunology, Children’s Hospital Boston, Boston, MA, USA
Talal Chatila - Laboratory of Neuroinflammation and Neurogenetics, Necker Branch, INSERM UMR1163, Paris Descartes University, Sorbonne-Paris-Cité, Institut Imagine, Paris, France
Yanick J. Crow - Division of Evolution and Genomic Sciences, School of Biological Sciences, Faculty of Biology, Medicine and Health, University of Manchester, Manchester Academic Health Science Centre, Manchester, UK
Yanick J. Crow - Departments of Medicine and Pediatrics, Mount Sinai School of Medicine, NewYork, NY, USA
Charlotte Cunningham-Rundles - Ruth’s Children’s Hospital-Technion, Haifa, Israel
Amos Etzioni - Grupo de Inmunodeficiencias Primarias, Facultad de Medicina, Universidad de Antioquia UdeA, Medellin, Colombia
Jose Luis Franco - Laboratory of Clinical Infectious Diseases, National Institute of Allergy and Infectious Diseases, Bethesda, MD, USA
Steven M. Holland - Dr von Hauner Children’s Hospital, Ludwig-Maximilians-University Munich, Munich, Germany
Christoph Klein - Department of Pediatrics and Developmental Biology, Tokyo Medical and Dental University (TMDU), Tokyo, Japan
Tomohiro Morio - Department of Pediatrics, University of Washington and Seattle Children’s Research Institute, Seattle, WA, USA
Hans D. Ochs & Troy R. Torgerson - Department of Clinical Immunology, Hôpital Saint-Louis, Assistance Publique-Hôpitaux de Paris, University Paris Diderot, Sorbonne Paris Cité, Paris, France
Eric Oksenhendler - Department of Pediatrics, University of California San Francisco and UCSF Benioff Children’s Hospital, San Francisco, CA, USA
Jennifer Puck - Murdoch Children’s Research Institute, Melbourne, VIC, Australia
Mimi L. K. Tang - Department of Paediatrics, University of Melbourne, Melbourne, VIC, Australia
Mimi L. K. Tang - Department of Allergy and Immunology, Royal Children’s Hospital, Melbourne, Australia
Mimi L. K. Tang - Immunology Division, Garvan Institute of Medical Research, Darlinghurst, NSW, Australia
Stuart G. Tangye - St Vincent’s Clinical School, University of NSW, Sydney, Australia
Stuart G. Tangye - Division of Allergy Immunology, Department of Pediatrics, The Children’s Hospital of Philadelphia, University of Pennsylvania Perelman School of Medicine, ARC 1216-I 3615 Civic Center Blvd, Philadelphia, PA, 19104, USA
Kathleen E. Sullivan
Authors
- Capucine Picard
You can also search for this author inPubMed Google Scholar - H. Bobby Gaspar
You can also search for this author inPubMed Google Scholar - Waleed Al-Herz
You can also search for this author inPubMed Google Scholar - Aziz Bousfiha
You can also search for this author inPubMed Google Scholar - Jean-Laurent Casanova
You can also search for this author inPubMed Google Scholar - Talal Chatila
You can also search for this author inPubMed Google Scholar - Yanick J. Crow
You can also search for this author inPubMed Google Scholar - Charlotte Cunningham-Rundles
You can also search for this author inPubMed Google Scholar - Amos Etzioni
You can also search for this author inPubMed Google Scholar - Jose Luis Franco
You can also search for this author inPubMed Google Scholar - Steven M. Holland
You can also search for this author inPubMed Google Scholar - Christoph Klein
You can also search for this author inPubMed Google Scholar - Tomohiro Morio
You can also search for this author inPubMed Google Scholar - Hans D. Ochs
You can also search for this author inPubMed Google Scholar - Eric Oksenhendler
You can also search for this author inPubMed Google Scholar - Jennifer Puck
You can also search for this author inPubMed Google Scholar - Mimi L. K. Tang
You can also search for this author inPubMed Google Scholar - Stuart G. Tangye
You can also search for this author inPubMed Google Scholar - Troy R. Torgerson
You can also search for this author inPubMed Google Scholar - Kathleen E. Sullivan
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toKathleen E. Sullivan.
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Picard, C., Bobby Gaspar, H., Al-Herz, W. et al. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity.J Clin Immunol 38, 96–128 (2018). https://doi.org/10.1007/s10875-017-0464-9
- Received: 19 July 2017
- Accepted: 31 October 2017
- Published: 11 December 2017
- Issue Date: January 2018
- DOI: https://doi.org/10.1007/s10875-017-0464-9