Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism (original) (raw)
- Letter to the Editor
- Published: 13 November 2012
Molecular Psychiatry volume 18, pages 1054–1056 (2013)Cite this article
- 3281 Accesses
- 7 Altmetric
- Metrics details
Subjects
Four recent studies of the coding regions of the human genome (the ‘exome’), suggest that new (de novo) mutations in hundreds of genes may contribute to the risk of autism spectrum disorder (ASD).1, 2, 3, 4 While the experimental strategy in the different efforts is almost identical, the four studies were published independently, and no integrative analysis has yet been reported. Notably, limited conclusions regarding the specific systems of genes disrupted by de novo mutations can be drawn based on each study alone. This stems from the relatively small fraction of mutations identified in each study in which there is a clear functional phenotype at the protein level. Here we show that upon combining the evidence from the different studies and integrating it with gene expression data from the developing human brain, a large group of genes emerges that is involved in regulation of expression during prenatal brain development. This suggests a prominent role in ASD for the genes involved in transcription regulation during brain development, specifically chromatin regulators.
We analyzed the complete collection of de novo single-nucleotide variations (SNVs) that were identified in 965 probands, sequenced in the four studies.1, 2, 3, 4 We gathered 121 genes that are most likely to be disrupted, containing de novo SNVs, which are nonsense, frameshift or splice site mutations. To characterize the genes, we first analyzed the enrichment of cellular processes and gene ontology (GO), using the Database for Annotation, Visualization and Integrated Discovery (DAVID). We found a significant enrichment for ‘chromatin regulator’ (UniProtKB; corrected _P_=0.021; Enrichment Score=2.04). Among the genes in this category, the chromatin remodeling gene CHD8 harbored two mutations in separate individuals (a frameshift and a nonsense mutation).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
Figure 1
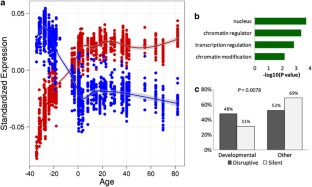
References
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J et al. De novo gene disruptions in children on the autistic spectrum. Neuron 2012; 74: 285–299.
Article CAS PubMed PubMed Central Google Scholar - Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 2012; 485: 242–245.
Article CAS PubMed PubMed Central Google Scholar - O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012; 485: 246–250.
Article PubMed PubMed Central Google Scholar - Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012; 485: 237–241.
Article CAS PubMed PubMed Central Google Scholar - Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 2012; 337: 64–69.
Article CAS PubMed PubMed Central Google Scholar - Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M et al. Spatio-temporal transcriptome of the human brain. Nature 2011; 478: 483–489.
Article CAS PubMed PubMed Central Google Scholar - Yoo AS, Crabtree GR . ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol 2009; 19: 120–126.
Article CAS PubMed PubMed Central Google Scholar - Kleefstra T, Kramer JM, Neveling K, Willemsen MH, Koemans TS, Vissers LE et al. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet 2012; 91: 73–82.
Article CAS PubMed PubMed Central Google Scholar
Acknowledgements
This research was supported by a grant from the National Institute for Psychobiology in Israel.
Author information
Authors and Affiliations
- Department of Genetics, The Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem, Israel
E Ben-David & S Shifman
Authors
- E Ben-David
You can also search for this author inPubMed Google Scholar - S Shifman
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toS Shifman.
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
Ben-David, E., Shifman, S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism.Mol Psychiatry 18, 1054–1056 (2013). https://doi.org/10.1038/mp.2012.148
- Published: 13 November 2012
- Issue Date: October 2013
- DOI: https://doi.org/10.1038/mp.2012.148