Genome sequencing in microfabricated high-density picolitre reactors (original) (raw)
Main
DNA sequencing has markedly changed the nature of biomedical research and medicine. Reductions in the cost, complexity and time required to sequence large amounts of DNA, including improvements in the ability to sequence bacterial and eukaryotic genomes, will have significant scientific, economic and cultural impact. Large-scale sequencing projects, including whole-genome sequencing, have usually required the cloning of DNA fragments into bacterial vectors, amplification and purification of individual templates, followed by Sanger sequencing1 using fluorescent chain-terminating nucleotide analogues2 and either slab gel or capillary electrophoresis. Current estimates put the cost of sequencing a human genome between 10millionand10 million and 10millionand25 million[3](/articles/nature03959#ref-CR3 "NIH News Release. NHGRI seeks next generation of sequencing technologies. 14 October 2004 http://www.genome.gov/12513210
."). Alternative sequencing methods have been described[4](/articles/nature03959#ref-CR4 "Nyren, P., Pettersson, B. & Uhlen, M. Solid phase DNA minisequencing by an enzymatic luminometric inorganic pyrophosphate detection assay. Anal. Biochem. 208, 171–175 (1993)"),[5](/articles/nature03959#ref-CR5 "Ronaghi, M. et al. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 242, 84–89 (1996)"),[6](/articles/nature03959#ref-CR6 "Jacobson, K. B. et al. Applications of mass spectrometry to DNA sequencing. GATA 8, 223–229 (1991)"),[7](/articles/nature03959#ref-CR7 "Bains, W. & Smith, G. C. A novel method for nucleic acid sequence determination. J. Theor. Biol. 135, 303–307 (1988)"),[8](/articles/nature03959#ref-CR8 "Jett, J. H. et al. High-speed DNA sequencing: an approach based upon fluorescence detection of single molecules. Biomol. Struct. Dynam. 7, 301–309 (1989)"); however, no technology has displaced the use of bacterial vectors and Sanger sequencing as the main generators of sequence information.Here we describe an integrated system whose throughput routinely enables applications requiring millions of bases of sequence information, including whole-genome sequencing. Our focus has been on the co-development of an emulsion-based method9,10,11 to isolate and amplify DNA fragments in vitro, and of a fabricated substrate and instrument that performs pyrophosphate-based sequencing (pyrosequencing5,12) in picolitre-sized wells.
In a typical run we generate over 25 million bases with a Phred quality score of 20 or better (predicted to have an accuracy of 99% or higher). Although this Phred 20 quality throughput is significantly higher than that of Sanger sequencing by capillary electrophoresis, it is currently at the cost of substantially shorter reads and lower average individual read accuracy. Sanger-based capillary electrophoresis sequencing systems produce up to 700 bases of sequence information from each of 96 DNA templates at an average read accuracy of 99.4% in 1 h, or 67,000 bases per hour, with substantially all of the bases having Phred 20 or better quality23. We further characterize the performance of the system and demonstrate that it is possible to assemble bacterial genomes de novo from relatively short reads by sequencing a known bacterial genome, Mycoplasma genitalium (580,069 bases), and comparing our shotgun sequencing and de novo assembly with the results originally obtained for this genome13. The results of shotgun sequencing and de novo assembly of a larger bacterial genome, that of Streptococcus pneumoniae14 (2.1 megabases (Mb)), are presented in Supplementary Table 4.
Emulsion-based sample preparation
We generate random libraries of DNA fragments by shearing an entire genome and isolating single DNA molecules by limiting dilution (Supplementary Methods). Specifically, we randomly fragment the entire genome, add specialized common adapters to the fragments, capture the individual fragments on their own beads and, within the droplets of an emulsion, clonally amplify the individual fragment (Fig. 1a, b). Unlike in current sequencing technology, our approach does not require subcloning in bacteria or the handling of individual clones; the templates are handled in bulk within the emulsions9,10,11.
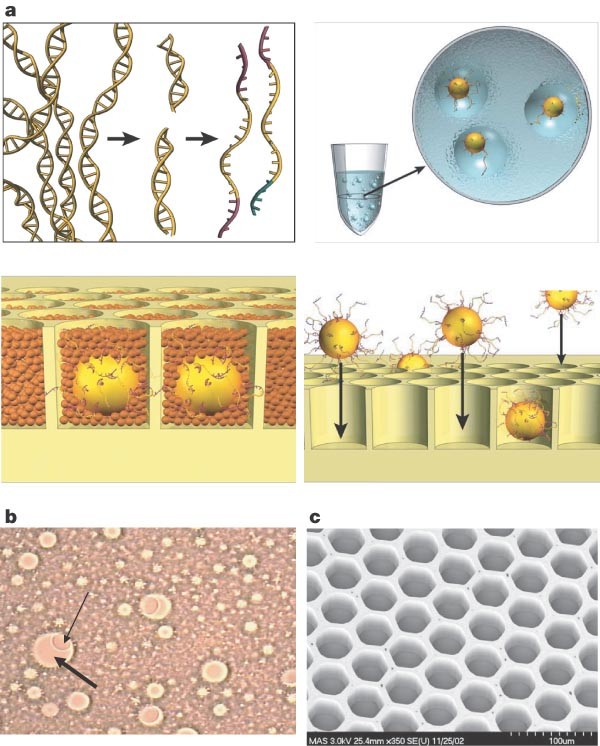
Figure 1: Sample preparation.
a, Genomic DNA is isolated, fragmented, ligated to adapters and separated into single strands (top left). Fragments are bound to beads under conditions that favour one fragment per bead, the beads are captured in the droplets of a PCR-reaction-mixture-in-oil emulsion and PCR amplification occurs within each droplet, resulting in beads each carrying ten million copies of a unique DNA template (top right). The emulsion is broken, the DNA strands are denatured, and beads carrying single-stranded DNA clones are deposited into wells of a fibre-optic slide (bottom right). Smaller beads carrying immobilized enzymes required for pyrophosphate sequencing are deposited into each well (bottom left). b, Microscope photograph of emulsion showing droplets containing a bead and empty droplets. The thin arrow points to a 28-µm bead; the thick arrow points to an approximately 100-µm droplet. c, Scanning electron micrograph of a portion of a fibre-optic slide, showing fibre-optic cladding and wells before bead deposition.
Sequencing in fabricated picolitre-sized reaction vessels
We perform sequencing by synthesis simultaneously in open wells of a fibre-optic slide using a modified pyrosequencing protocol that is designed to take advantage of the small scale of the wells. The fibre-optic slides are manufactured by slicing of a fibre-optic block that is obtained by repeated drawing and fusing of optic fibres. At each iteration, the diameters of the individual fibres decrease as they are hexagonally packed into bundles of increasing cross-sectional sizes. Each fibre-optic core is 44 µm in diameter and surrounded by 2–3 µm of cladding; etching of each core creates reaction wells approximately 55 µm in depth with a centre-to-centre distance of 50 µm (Fig. 1c), resulting in a calculated well size of 75 pl and a well density of 480 wells mm-2. The slide, containing approximately 1.6 million wells15, is loaded with beads and mounted in a flow chamber designed to create a 300-µm high channel, above the well openings, through which the sequencing reagents flow (Fig. 2a, b). The unetched base of the slide is in optical contact with a second fibre-optic imaging bundle bonded to a charge-coupled device (CCD) sensor, allowing the capture of emitted photons from the bottom of each individual well (Fig. 2c; see also Supplementary Methods).
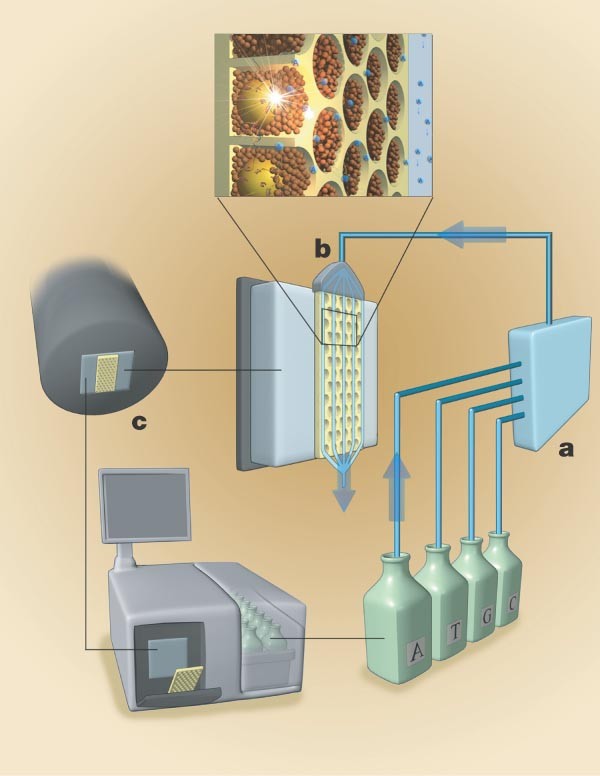
Figure 2: Sequencing instrument.
The sequencing instrument consists of the following major subsystems: a fluidic assembly (a), a flow chamber that includes the well-containing fibre-optic slide (b), a CCD camera-based imaging assembly (c), and a computer that provides the necessary user interface and instrument control.
We developed a three-bead system, and optimized the components to achieve high efficiency on solid support. The combination of picolitre-sized wells, enzyme loading uniformity allowed by the small beads and enhanced solid support chemistry enabled us to develop a method that extends the useful read length of sequencing-by-synthesis to 100 bases (Supplementary Methods).
In the flow chamber cyclically delivered reagents flow perpendicularly to the wells. This configuration allows simultaneous extension reactions on template-carrying beads within the open wells and relies on convective and diffusive transport to control the addition or removal of reagents and by-products. The timescale for diffusion into and out of the wells is on the order of 10 s in the current configuration and is dependent on well depth and flow channel height. The timescales for the signal-generating enzymatic reactions are on the order of 0.02–1.5 s (Supplementary Methods). The current reaction is dominated by mass transport effects, and improvements based on faster delivery of reagents are possible. Well depth was selected on the basis of a number of competing requirements: (1) wells need to be deep enough for the DNA-carrying beads to remain in the wells in the presence of convective transport past the wells; (2) they must be sufficiently deep to provide adequate isolation against diffusion of by-products from a well in which incorporation is taking place to a well where no incorporation is occurring; and (3) they must be shallow enough to allow rapid diffusion of nucleotides into the wells and rapid washing out of remaining nucleotides at the end of each flow cycle to enable high sequencing throughput and reduced reagent use. After the flow of each nucleotide, a wash containing apyrase is used to ensure that nucleotides do not remain in any well before the next nucleotide being introduced.
Base calling of individual reads
Nucleotide incorporation is detected by the associated release of inorganic pyrophosphate and the generation of photons5,12. Wells containing template-carrying beads are identified by detecting a known four-nucleotide ‘key’ sequence at the beginning of the read (Supplementary Methods). Raw signals are background-subtracted, normalized and corrected. The normalized signal intensity at each nucleotide flow, for a particular well, indicates the number of nucleotides, if any, that were incorporated. This linearity in signal is preserved to at least homopolymers of length eight (Supplementary Fig. 6). In sequencing by synthesis a very small number of templates on each bead lose synchronism (that is, either get ahead of, or fall behind, all other templates in sequence16). The effect is primarily due to leftover nucleotides in a well (creating ‘carry forward’) or to incomplete extension. Typically, we observe a carry forward rate of 1–2% and an incomplete extension rate of 0.1–0.3%. Correction of these shifts is essential because the loss of synchronism is a cumulative effect that degrades the quality of sequencing at longer read lengths. We have developed algorithms, based on detailed models of the underlying physical phenomena, that allow us to determine, and correct for, the amounts of carry forward and incomplete extension occurring in individual wells (Supplementary Methods). Figure 3 shows the processed result, a 113-bases-long read generated in the M. genitalium run discussed below. To assess sequencing performance and the effectiveness of the correction algorithms, independently of artefacts introduced during the emulsion-based sample preparation, we created test fragments with difficult-to-sequence stretches of identical bases of increasing length (homopolymers) (Supplementary Methods and Supplementary Fig. 4). Using these test fragments, we have verified that at the individual read level we achieve base call accuracy of approximately 99.4%, at read lengths in excess of 100 bases (Table 1).
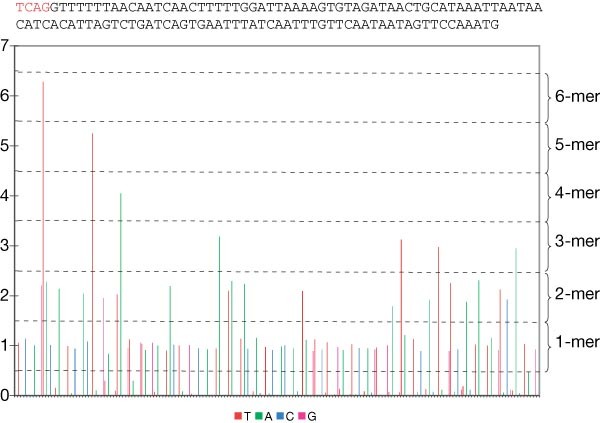
Figure 3: Flowgram of a 113-bases read from an M. genitalium run.
Nucleotides are flowed in the order T, A, C, G. The sequence is shown above the flowgram. The signal value intervals corresponding to the various homopolymers are indicated on the right. The first four bases (in red, above the flowgram) constitute the ‘key’ sequence, used to identify wells containing a DNA-carrying bead.
Table 1 Summary of sequencing statistics for test fragments
High-quality reads and consensus accuracy
Before base calling or aligning reads, we select high-quality reads without relying on a priori knowledge of the genome or template being sequenced (Supplementary Methods). This selection is based on the observation that poor-quality reads have a high proportion of signals that do not allow a clear distinction between a flow during which no nucleotide was incorporated and a flow during which one or more nucleotide was incorporated. When base calling individual reads, errors can occur because of signals that have ambiguous values (Supplementary Fig. 5). To improve the usability of our reads, we also developed a metric that allows us to estimate ab initio the quality (or probability of correct base call) of each base of a read, analogous to the Phred score17 used by current Sanger sequencers (Supplementary Methods and Supplementary Fig. 8).
Higher quality sequence can be achieved by taking advantage of the high over sampling that our system affords and building a consensus sequence. Sequences are aligned to one another using the signal strengths at each nucleotide flow, rather than individual base calls, to determine optimal alignment (Supplementary Methods). The corresponding signals are then averaged, after which base calling is performed. This approach greatly improves the accuracy of the sequence (Supplementary Fig. 7) and provides an estimate of the quality of the consensus base. We refer to that quality measure as the _Z_-score—it is a measure of the spread of signals in all the reads at one location and the distance between the average signal and the closest base-calling threshold value. In both re-sequencing and de novo sequencing, as the minimum _Z_-score is raised the consensus accuracy increases, while coverage decreases; approximately half of the excluded bases, as the _Z_-score is increased, belong to homopolymers of length four and larger. Sanger sequencers usually require a depth of coverage at any base of three or more in order to achieve a consensus accuracy of 99.99%. To achieve a minimum of threefold coverage of 95% of the unique portions of a typical genome requires approximately seven- to eightfold over sampling. Owing to our higher error rate, we have observed that comparable consensus accuracies, over a similar fraction of a genome, are achieved with a depth of coverage of four or more, requiring approximately ten to twelve times over sampling.
Mycoplasma genitalium
Mycoplasma genomic DNA was fragmented and prepared into a sequencing library as described above. (This was accomplished by a single individual in 4 h.) After emulsion polymerase chain reaction (PCR) and bead deposition onto a 60 × 60 mm2 fibre-optic slide, a process which took one individual 6 h, 42 cycles of four nucleotides were flowed through the sequencing system in an automated 4-h run of the instrument. The results are summarized in Table 2. In order to measure the quality of individual reads, we aligned each high quality read to the reference genome at 70% stringency using flow-space mapping and criteria similar to those used previously in assessing the accuracy of other base callers17. When assessing sequencing quality, only reads that mapped to unique locations in the reference genome were included. Because this process excludes repeat regions (parts of the genome for which corresponding flowgrams are 70% similar to one another), the selected reads did not cover the genome completely. Figure 4a illustrates the distribution of read lengths for this run. The average read length was 110 bases, the resulting over sample 40-fold, and 84,011 reads (27.4%) were perfect. Figure 4b summarizes the average error as a function of base position. Coverage of non-repeat regions was consistent with the sample preparation and emulsion not being biased (Supplementary Fig. 8). At the individual read level, we observe an insertion and deletion error rate of approximately 3.3%; substitution errors have a much lower rate, on the order of 0.5%. When using these reads without any _Z_-score restriction, we covered 99.94% of the genome in ten contiguous regions with a consensus accuracy of 99.97%. The error rate in homopolymers is significantly reduced in the consensus sequence (Supplementary Fig. 7). Of the bases not covered by this consensus sequence (366 bases), all belonged to excluded repeat regions. Setting a minimum _Z_-score equal to 4, coverage was reduced to 98.1% of the genome, while consensus accuracy increased to 99.996%. We further demonstrated the reproducibility of the system by repeating the whole-genome sequencing of M. genitalium an additional eight times, achieving a 40-fold coverage of the genome in each of the eight separate instrument runs (Supplementary Table 3).
Table 2 Summary statistics for M. genitalium
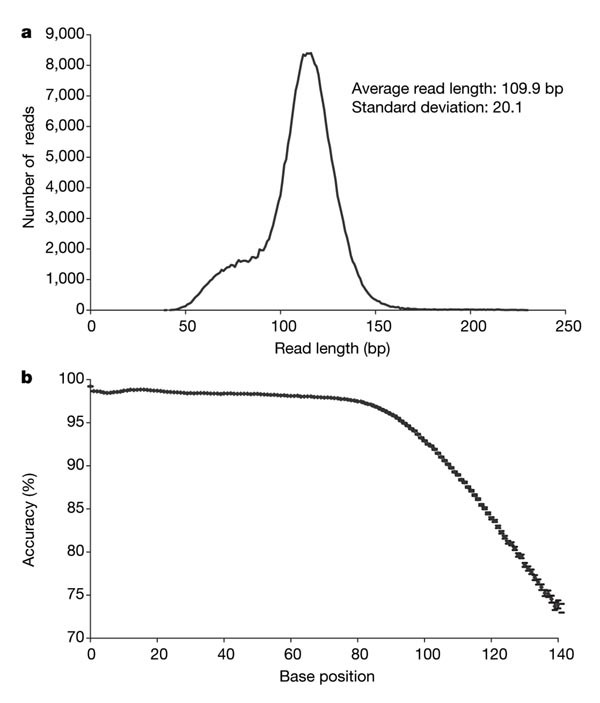
Figure 4: M. genitalium data.
a, Read length distribution for the 306,178 high-quality reads of the M. genitalium sequencing run. This distribution reflects the base composition of individual sequencing templates. b, Average read accuracy, at the single read level, as a function of base position for the 238,066 mapped reads of the same run.
We assembled the M. genitalium reads from a single run into 25 contigs with an average length of 22.4 kb. One of these contigs was misassembled due to a collapsed tandem repeat region of 60 bases, and was corrected by hand. The original sequencing of M. genitalium resulted in 28 contigs before directed sequencing used for finishing the sequence13. Our assembly covered 96.54% of the genome and attained a consensus accuracy of 99.96%. Non-resolvable repeat regions amount to 3% of the genome: we therefore covered 99.5% of the unique portions of the genome. Sixteen of the breaks between contigs were due to non-resolvable repeat regions, two were due to missed overlapping reads (our read filter and trimmer are not perfect and the algorithms we use to perform the pattern matching of flowgrams occasionally miss valid overlaps) and the remainder to thin read coverage. Setting a minimum _Z_-score of 4, coverage was reduced to 95.27% of the genome (98.2% of the resolvable part of the genome) with the consensus accuracy increasing to 99.994%.
Discussion
We have demonstrated the simultaneous acquisition of hundreds of thousands of sequence reads, 80–120 bases long, at 96% average accuracy in a single run of the instrument using a newly developed in vitro sample preparation methodology and sequencing technology. With Phred 20 as a cutoff, we show that our instrument is able to produce over 47 million bases from test fragments and 25 million bases from genomic libraries. We used test fragments to de-couple our sample preparation methodology from our sequencing technology. The decrease in single-read accuracy from 99.4% for test fragments to 96% for genomic libraries is primarily due to a lack of clonality in a fraction of the genomic templates in the emulsion, and is not an inherent limitation of the sequencing technology. Most of the remaining errors result from a broadening of signal distributions, particularly for large homopolymers (seven or more), leading to ambiguous base calls. Recent work on the sequencing chemistry and algorithms that correct for crosstalk between wells suggests that the signal distributions will narrow, with an attendant reduction in errors and increase in read lengths. In preliminary experiments with genomic libraries that also include improvements in the emulsion protocol, we are able to achieve, using 84 cycles, read lengths of 200 bases with accuracies similar to those demonstrated here for 100 bases. On occasion, at 168 cycles, we have generated individual reads that are 100% accurate over greater then 400 bases.
Using M. genitalium we demonstrate that short fragments a priori do not prohibit the de novo assembly of bacterial genomes. In fact, the larger over sampling afforded by the throughput of our system resulted in a draft sequence having fewer contigs than with Sanger reads, with substantially less effort. By taking advantage of the over sampling, consensus accuracies greater then 99.96% were achieved for this genome. Further quality filtering of the assembly results in the selection of a consensus sequence with accuracy exceeding 99.99% while incurring only a minor loss of genome coverage. Comparable results were seen when we shotgun sequenced and de novo assembled the 2.1-Mb genome of Streptococcus pneumoniae14 (Supplementary Table 4). The de novo assembly of genomes more complex than bacteria, including mammalian genomes, may require the development of methods similar to those developed for Sanger sequencing, to prepare and sequence paired end libraries that can span repeats in these genome. To facilitate the use of paired end libraries we have developed methods to sequence, in an individual well, from both ends of genomic template, and plan to add paired end read capabilities to our assembler (Supplementary Methods).
Future increases in throughput, and a concomitant reduction in cost per base, may come from the continued miniaturization of the fibre-optic reactors, allowing more sequence to be produced per unit area—a scaling characteristic similar to that which enabled the prediction of significant improvements in the integrated circuit at the start of its development cycle18.
Methods
Emulsion-based clonal amplification
The simultaneous amplification of fragments is achieved by isolating individual DNA-carrying beads in separate ∼100-µm aqueous droplets (on the order of 2 × 106 ml-1) made through the creation of a PCR-reaction-mixture-in-oil emulsion. (Fig. 1b; see also Supplementary Methods). The droplets act as separate microreactors in which parallel DNA amplifications are performed, yielding approximately 107 copies of a template per bead; 800 µl of emulsion containing 1.5 million beads are prepared in a standard 2-ml tube. Each emulsion is aliquoted into eight PCR tubes for amplification. After PCR, the emulsion is broken to release the beads, which include beads with amplified, immobilized DNA template and empty beads (Supplementary Methods). We then enrich for template-carrying beads (Supplementary Methods). Typically, about 30% of the beads will have DNA, producing 450,000 template-carrying beads per emulsion reaction. The number of emulsions prepared depends on the size of the genome and the expected number of runs required to achieve adequate over sampling. The 580-kb M. genitalium genome, sequenced on one 60 × 60 mm2 fibre-optic slide, required 1.6 ml of emulsion. A human genome, over sampled ten times, would require approximately 3,000 ml of emulsion.
Bead loading into picolitre wells
The enriched template-carrying beads are deposited by centrifugation into open wells (Fig. 1c), arranged along one face of a 60 × 60 mm2 fibre-optic slide. The beads (diameter ∼28 µm) are sized to ensure that no more than one bead fits in most wells (we observed that 2–5% of filled wells contain more than one bead). Loading 450,000 beads (from one emulsion preparation) onto each half of a 60 × 60 mm2 plate was experimentally found to limit bead occupancy to approximately 35% of all wells, thereby reducing chemical and optical crosstalk between wells. A mixture of smaller beads that carry immobilized ATP sulphurylase and luciferase necessary to generate light from free pyrophosphate are also loaded into the wells to create the individual sequencing reactors (Supplementary Methods).
Image capture
A bead carrying 10 million copies of a template yields approximately 10,000 photons at the CCD sensor, per incorporated nucleotide. The generated light is transmitted through the base of the fibre-optic slide and detected by a large format CCD (4,095 × 4,096 pixels). The images are processed to yield sequence information simultaneously for all wells containing template-carrying beads. The imaging system was designed to accommodate a large number of small wells and the large number of optical signals being generated from individual wells during each nucleotide flow. Once mounted, the fibre-optic slide's position does not shift; this makes it possible for the image analysis software to determine the location of each well (whether or not it contains a DNA-carrying bead), based on light generation during the flow of a pyrophosphate solution, which precedes each sequencing run. A single well is imaged by approximately nine 15 µm pixels. For each nucleotide flow, the light intensities collected by the pixels covering a particular well are summed to generate a signal for that particular well at that particular nucleotide flow. Each image captured by the CCD produces 32 megabytes of data. In order to perform all of the necessary signal processing in real time, the control computer is fitted with an accessory board (Supplementary Methods), hosting a 6 million gate Field Programmable Gate Array (FPGA)19,20.
De novo shotgun sequence assembler
A de novo flow-space assembler was developed to capture all of the information contained in the original flow-based signal trace. It also addresses the fact that existing assemblers are not optimized for 80–120-bases reads, particularly with respect to memory management due to the increased number of sequencing reads needed to achieve equivalent genome coverage. (A completely random genome covered with 100-bases reads requires approximately 50% more reads to yield the same number of contiguous regions (contigs) as achieved with 700-bases reads, assuming the need for a 30-bases overlap between reads21.) This assembler consists of a series of modules: the Overlapper, which finds and creates overlaps between reads; the Unitigger, which constructs larger contigs of overlapping sequence reads; and the Multialigner, which generates consensus calls and quality scores for the bases within each contig (Supplementary Methods). (The names of the software modules are based on those performing related functions in other assemblers developed previously22.)
References
- Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA 74, 5463–5467 (1977)
ADS CAS PubMed PubMed Central Google Scholar - Prober, J. M. et al. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science 238, 336–341 (1987)
Article ADS CAS PubMed Google Scholar - NIH News Release. NHGRI seeks next generation of sequencing technologies. 14 October 2004 http://www.genome.gov/12513210.
- Nyren, P., Pettersson, B. & Uhlen, M. Solid phase DNA minisequencing by an enzymatic luminometric inorganic pyrophosphate detection assay. Anal. Biochem. 208, 171–175 (1993)
Article CAS PubMed Google Scholar - Ronaghi, M. et al. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 242, 84–89 (1996)
Article CAS PubMed Google Scholar - Jacobson, K. B. et al. Applications of mass spectrometry to DNA sequencing. GATA 8, 223–229 (1991)
CAS Google Scholar - Bains, W. & Smith, G. C. A novel method for nucleic acid sequence determination. J. Theor. Biol. 135, 303–307 (1988)
Article CAS PubMed Google Scholar - Jett, J. H. et al. High-speed DNA sequencing: an approach based upon fluorescence detection of single molecules. Biomol. Struct. Dynam. 7, 301–309 (1989)
Article CAS Google Scholar - Tawfik, D. S. & Griffiths, A. D. Man-made cell-like compartments for molecular evolution. Nature Biotechnol. 16, 652–656 (1998)
Article CAS Google Scholar - Ghadessy, F. J., Ong, J. L. & Holliger, P. Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl Acad. Sci. USA 98, 4552–4557 (2001)
Article ADS CAS PubMed PubMed Central Google Scholar - Dressman, D., Yan, H., Traverso, G., Kinzler, K. W. & Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA 100, 8817–8822 (2003)
Article ADS CAS PubMed PubMed Central Google Scholar - Ronaghi, M., Uhlen, M. & Nyren, P. A sequencing method based on real-time pyrophosphate. Science 281, 363–365 (1998)
Article CAS PubMed Google Scholar - Fraser, C. M. et al. The minimal gene complement of Mycoplasma genitalium. Science 270, 397–403 (1995)
Article ADS CAS PubMed Google Scholar - Tettelin, H. et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506 (2001)
Article CAS PubMed Google Scholar - Leamon, J. H. et al. A massively parallel PicoTiterPlate based platform for discrete picoliter-scale polymerase chain reactions. Electrophoresis 24, 3769–3777 (2003)
Article CAS PubMed Google Scholar - Ronaghi, M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 11, 3–11 (2001)
Article CAS PubMed Google Scholar - Ewing, B., Hillier, L., Wendl, M. C. & Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185 (1998)
CAS PubMed Google Scholar - Moore, G. E. Cramming more components onto integrated circuits. Electronics 38(8) (1965)
- Mehta, K., Rajesh, V. A. & Veeraswamy, S. FPGA implementation of VXIbus interface hardware. Biomed. Sci. Instrum. 29, 507–513 (1993)
CAS PubMed Google Scholar - Fagin, B., Watt, J. G. & Gross, R. A special-purpose processor for gene sequence analysis. Comput. Appl. Biosci. 9, 221–226 (1993)
CAS PubMed Google Scholar - Lander, E. S. & Waterman, M. S. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2, 231–239 (1988)
Article CAS PubMed Google Scholar - Myers, E. W. Toward simplifying and accurately formulating fragment assembly. J Comput. Biol. 2, 275–290 (1995)
Article CAS PubMed Google Scholar - Ogawa, T. et al. Increased Productivity For Core Labs Using One Polymer and One Array Length for Multiple Applications. Poster P108-T. ABRF '05: Biomolecular Technologies: Discovery to Hypotheses (Savannah, Georgia, 5–8 February 2005); also available as Applied Biosystems 3730x/DNA Analyzer Specification Sheet (2004).
Acknowledgements
We acknowledge P. Dacey and the support of the Operations groups of 454 Life Sciences. This research was supported in part by the US Department of Health and Human Services under NIH grants.
Author information
Author notes
- Marcel Margulies and Michael Egholm: *These authors contributed equally to this work
Authors and Affiliations
- 454 Life Sciences Corp., 20 Commercial Street, Connecticut, 06405, Branford, USA
Marcel Margulies, Michael Egholm, William E. Altman, Said Attiya, Joel S. Bader, Lisa A. Bemben, Jan Berka, Michael S. Braverman, Yi-Ju Chen, Zhoutao Chen, Scott B. Dewell, Lei Du, Joseph M. Fierro, Xavier V. Gomes, Brian C. Godwin, Wen He, Scott Helgesen, Chun He Ho, Gerard P. Irzyk, Szilveszter C. Jando, Maria L. I. Alenquer, Thomas P. Jarvie, Kshama B. Jirage, Jong-Bum Kim, James R. Knight, Janna R. Lanza, John H. Leamon, Steven M. Lefkowitz, Ming Lei, Jing Li, Kenton L. Lohman, Hong Lu, Vinod B. Makhijani, Keith E. McDade, Michael P. McKenna, Elizabeth Nickerson, John R. Nobile, Ramona Plant, Bernard P. Puc, Michael T. Ronan, George T. Roth, Gary J. Sarkis, Jan Fredrik Simons, John W. Simpson, Maithreyan Srinivasan, Karrie R. Tartaro, Kari A. Vogt, Greg A. Volkmer, Shally H. Wang, Yong Wang, Pengguang Yu, Richard F. Begley & Jonathan M. Rothberg - University of California, California, 94720, Berkeley, USA
Eugene W. Myers - Laboratory of Microbiology, The Rockefeller University, New York, 10021, New York, USA
Alexander Tomasz - The Rothberg Institute for Childhood Diseases, 530 Whitfield Street, Connecticut, 06437, Guilford, USA
Michael P. Weiner
Authors
- Marcel Margulies
You can also search for this author inPubMed Google Scholar - Michael Egholm
You can also search for this author inPubMed Google Scholar - William E. Altman
You can also search for this author inPubMed Google Scholar - Said Attiya
You can also search for this author inPubMed Google Scholar - Joel S. Bader
You can also search for this author inPubMed Google Scholar - Lisa A. Bemben
You can also search for this author inPubMed Google Scholar - Jan Berka
You can also search for this author inPubMed Google Scholar - Michael S. Braverman
You can also search for this author inPubMed Google Scholar - Yi-Ju Chen
You can also search for this author inPubMed Google Scholar - Zhoutao Chen
You can also search for this author inPubMed Google Scholar - Scott B. Dewell
You can also search for this author inPubMed Google Scholar - Lei Du
You can also search for this author inPubMed Google Scholar - Joseph M. Fierro
You can also search for this author inPubMed Google Scholar - Xavier V. Gomes
You can also search for this author inPubMed Google Scholar - Brian C. Godwin
You can also search for this author inPubMed Google Scholar - Wen He
You can also search for this author inPubMed Google Scholar - Scott Helgesen
You can also search for this author inPubMed Google Scholar - Chun He Ho
You can also search for this author inPubMed Google Scholar - Gerard P. Irzyk
You can also search for this author inPubMed Google Scholar - Szilveszter C. Jando
You can also search for this author inPubMed Google Scholar - Maria L. I. Alenquer
You can also search for this author inPubMed Google Scholar - Thomas P. Jarvie
You can also search for this author inPubMed Google Scholar - Kshama B. Jirage
You can also search for this author inPubMed Google Scholar - Jong-Bum Kim
You can also search for this author inPubMed Google Scholar - James R. Knight
You can also search for this author inPubMed Google Scholar - Janna R. Lanza
You can also search for this author inPubMed Google Scholar - John H. Leamon
You can also search for this author inPubMed Google Scholar - Steven M. Lefkowitz
You can also search for this author inPubMed Google Scholar - Ming Lei
You can also search for this author inPubMed Google Scholar - Jing Li
You can also search for this author inPubMed Google Scholar - Kenton L. Lohman
You can also search for this author inPubMed Google Scholar - Hong Lu
You can also search for this author inPubMed Google Scholar - Vinod B. Makhijani
You can also search for this author inPubMed Google Scholar - Keith E. McDade
You can also search for this author inPubMed Google Scholar - Michael P. McKenna
You can also search for this author inPubMed Google Scholar - Eugene W. Myers
You can also search for this author inPubMed Google Scholar - Elizabeth Nickerson
You can also search for this author inPubMed Google Scholar - John R. Nobile
You can also search for this author inPubMed Google Scholar - Ramona Plant
You can also search for this author inPubMed Google Scholar - Bernard P. Puc
You can also search for this author inPubMed Google Scholar - Michael T. Ronan
You can also search for this author inPubMed Google Scholar - George T. Roth
You can also search for this author inPubMed Google Scholar - Gary J. Sarkis
You can also search for this author inPubMed Google Scholar - Jan Fredrik Simons
You can also search for this author inPubMed Google Scholar - John W. Simpson
You can also search for this author inPubMed Google Scholar - Maithreyan Srinivasan
You can also search for this author inPubMed Google Scholar - Karrie R. Tartaro
You can also search for this author inPubMed Google Scholar - Alexander Tomasz
You can also search for this author inPubMed Google Scholar - Kari A. Vogt
You can also search for this author inPubMed Google Scholar - Greg A. Volkmer
You can also search for this author inPubMed Google Scholar - Shally H. Wang
You can also search for this author inPubMed Google Scholar - Yong Wang
You can also search for this author inPubMed Google Scholar - Michael P. Weiner
You can also search for this author inPubMed Google Scholar - Pengguang Yu
You can also search for this author inPubMed Google Scholar - Richard F. Begley
You can also search for this author inPubMed Google Scholar - Jonathan M. Rothberg
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toJonathan M. Rothberg.
Ethics declarations
Competing interests
The authors declare employment and personal financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures S1-S11 that illustrate various experimental, data processing and modelling results pertinent to the sequencing technology described in the paper. (DOC 305 kb)
Supplementary Tables
This file includes Supplementary Tables S1-S4 with model results and sequencing statistics for a number of runs performed with the instrument described in the paper. (DOC 87 kb)
Supplementary Methods
This file includes detailed methods and materials for the sequencing technology described in the paper, and detailed description of data processing and bioinformatics algorithms. Earlier errors (see Corrigendum in Nature 441, page 120; 4 May 2006) have been corrected and are shown in red. (DOC 158 kb)
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/), which permits distribution, and reproduction in any medium, provided the original author and source are credited. This licence does not permit commercial exploitation, and derivative works must be licensed under the same or similar licence.
About this article
Cite this article
Margulies, M., Egholm, M., Altman, W. et al. Genome sequencing in microfabricated high-density picolitre reactors.Nature 437, 376–380 (2005). https://doi.org/10.1038/nature03959
- Received: 06 May 2005
- Accepted: 10 June 2005
- Published: 31 July 2005
- Issue Date: 15 September 2005
- DOI: https://doi.org/10.1038/nature03959