Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans (original) (raw)
Abstract
Phytophthora infestans is the most destructive pathogen of potato and a model organism for the oomycetes, a distinct lineage of fungus-like eukaryotes that are related to organisms such as brown algae and diatoms. As the agent of the Irish potato famine in the mid-nineteenth century, P. infestans has had a tremendous effect on human history, resulting in famine and population displacement1. To this day, it affects world agriculture by causing the most destructive disease of potato, the fourth largest food crop and a critical alternative to the major cereal crops for feeding the world’s population1. Current annual worldwide potato crop losses due to late blight are conservatively estimated at $6.7 billion2. Management of this devastating pathogen is challenged by its remarkable speed of adaptation to control strategies such as genetically resistant cultivars3,4. Here we report the sequence of the P. infestans genome, which at ∼240 megabases (Mb) is by far the largest and most complex genome sequenced so far in the chromalveolates. Its expansion results from a proliferation of repetitive DNA accounting for ∼74% of the genome. Comparison with two other Phytophthora genomes showed rapid turnover and extensive expansion of specific families of secreted disease effector proteins, including many genes that are induced during infection or are predicted to have activities that alter host physiology. These fast-evolving effector genes are localized to highly dynamic and expanded regions of the P. infestans genome. This probably plays a crucial part in the rapid adaptability of the pathogen to host plants and underpins its evolutionary potential.
Similar content being viewed by others
Main
The size of the P. infestans genome is estimated by optical map and other methods at 240 Mb (Supplementary Information). It is several-fold larger than those of the related Phytophthora species P. sojae (95 Mb) and P. ramorum (65 Mb), which cause soybean root rot and sudden oak death, respectively5,6. We sequenced the genome of P. infestans strain T30-4 using a whole-genome shotgun approach, and generated a ninefold coverage assembly spanning 229 Mb (Table 1 and Supplementary Information). The unassembled fraction of the genome consists of high copy repeat sequences (Supplementary Information). The assembled genome sequence provides near complete coverage of genes, with 98.2% of P. infestans T30-4 complementary DNAs aligning (Supplementary Information). We identified 17,797 protein-coding genes by ab initio gene prediction, protein and expressed sequence tag (EST) homology, and direct genome-to-genome comparative gene modelling with P. sojae and P. ramorum (Supplementary Information). Changes in gene content, number or length do not explain the marked difference in genome size (Table 1 and Supplementary Table 1). No evidence of whole-genome duplication or large-scale dispersed segmental duplication was detected. However, specific disease effector gene families are expanded in P. infestans (see later).
Table 1 Genome assembly and annotation statistics
P. infestans, P. sojae and P. ramorum represent three major phylogenetic clades of Phytophthora6. Among the three genomes, we identified a core set of 8,492 orthologue clusters (including 9,583 P. infestans orthologues and close paralogues), of which 7,113 genes show 1:1:1 orthology relationships (Table 1, Supplementary Fig. 1 and Supplementary Table 2). The core proteome is enriched in genes involved in cellular processes including DNA replication, transcription and protein translation, whereas genes with functions involved in cellular defence mechanisms are underrepresented (Supplementary Fig. 2). Differences in gene family expansion, in particular dynamic repertoires of effector genes (see later), are probably responsible for different traits among Phytophthora species, such as altered host specificity.
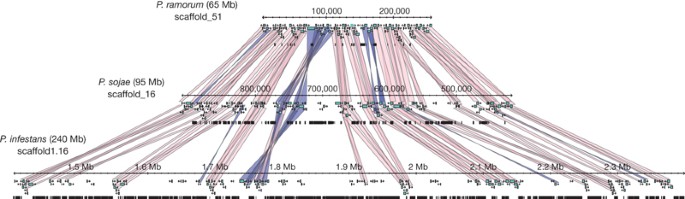
Comparison of the three Phytophthora genomes reveals an unusual genome organization, comprised of blocks of conserved gene order in which gene density is relatively high and repeat content is relatively low, separated by regions in which gene order is not conserved, gene density is low and repeat content is high (Table 1 and Fig. 1). The conserved blocks represent ∼90% of core orthologous groups in all three genomes, including ∼70% (12,440) of all P. infestans protein-coding genes and ∼78% of genes in both P. sojae (13,225) and P. ramorum (11,246). Within conserved blocks, genes are typically tightly spaced in all three genomes (Table 1 and Fig. 1), with median intergenic distances of 633 base pairs (bp) for P. ramorum, 804 bp for P. sojae, and 603 bp for P. infestans. In regions between conserved blocks, intergenic distances are greater and increase with increasing genome size (median 1.5 kb for P. ramorum, 2.2 kb for P. sojae, and 3.7 kb for P. infestans). The differences in spacing between genes among the three genomes, within and outside regions of conserved gene order, are evident in Fig. 2a–f. The expansion of regions between conserved blocks results from increased density of repetitive elements (Supplementary Fig. 3), and overall differences in genome size among the three species are largely explained by proliferation of repeats in regions in which gene order is not conserved. This difference between conserved blocks and non-conserved regions is particularly apparent in the greatly expanded P. infestans genome (Fig. 2d, f). Further, it is evident that rapidly evolving secreted effector genes (see later) lie predominantly in the gene-sparse regions (Fig. 2g, h). This dual pattern of intergenic spacing and repeat content has been suggested for large, unsequenced genomes in the Poaceae such as maize7,8,9, but it is not seen in the genomes of other sequenced eukaryotes (Supplementary Fig. 4).
Figure 1: Repeat-driven genome expansion in Phytophthora infestans.
Conserved gene order across three homologous Phytophthora scaffolds. Genome expansion is evident in regions of conserved gene order, a consequence of repeat expansion in intergenic regions. Genes are shown as turquoise boxes, repeats as black boxes. Collinear orthologous gene pairs are connected by pink (direct) or blue (inverted) bands.
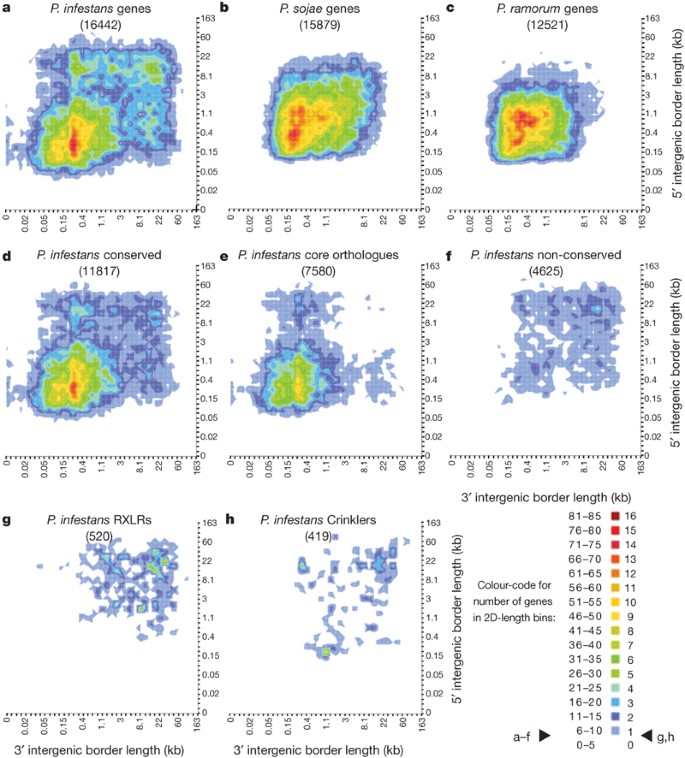
Figure 2: The P. infestans genome shows an unusual distribution of intergenic region lengths.
The flanking distance between neighbouring genes provides a measurement of local gene density. P. infestans genes were sorted into two dimensional bins on the basis of the lengths of flanking intergenic distances to neighbouring genes at their 5′ and 3′ ends. a–h, The number of genes in each bin is shown as a colour-coded heat map on orthogonal projection. P. infestans whole-genome analysis (a) shows most genes with intergenic regions between 20-bp and 3-kb long, as well as sets of genes flanked by one or two intergenic region(s) between 5 kb and 36 kb. Comparison with other Phytophthora genomes (b, c) indicates that this separation is observed in P. infestans but not the other two sequenced genomes. Genes in collinear blocks (d) and the core orthologue clusters (e) have primarily shorter intergenic distances, whereas genes outside of collinear blocks (f) reside mostly in gene sparse regions. Genes belonging to the RXLR (g) and Crinkler (CRN) (genes and pseudogenes) (h) effector families have flanking intergenic distances among the longest. Genes found at the ends of scaffolds and hence lacking neighbouring genes were necessarily excluded.
Recent proliferation of Gypsy elements in P. infestans underlies the genome expansion. Approximately one-third of the genome assembly corresponds to families of Gypsy elements (Supplementary Fig. 5). The two families with the highest relative expansion in P. infestans are Gypsy Pi-1 and a new Gypsy long terminal repeat (LTR) element we named ‘Albatross’, which together account for at least 29% of the genome (Supplementary Table 3). Albatross elements cover ∼32 Mb and are enriched (>2-fold) in the regions in which gene order is not conserved (Supplementary Table 4 and Supplementary Fig. 6), contributing appreciably to relative expansion of gene-sparse regions (Supplementary Fig. 3). Gypsy Pi-1 elements cover ∼22 Mb and, in contrast to Albatross elements, are relatively evenly distributed across the genome.
Overall, the P. infestans genome contains a strikingly rich and diverse population of transposons (Supplementary Table 3). We identified 273 full-length elements belonging to two large classes of autonomous rolling-circle type helitron DNA transposons (7.3-kb and 6.4-kb elements), in much larger numbers than described in any other genome (Supplementary Tables 3 and 5). Most helitron open reading frames (ORFs) are degenerate pseudogenes, but 13 are intact and presumed functional. Some apparently non-autonomous helitrons have intact termini so their transposition may be driven by gene products from the functional classes. In contrast, the P. sojae and P. ramorum genomes contain no intact helitron elements. The P. infestans genome carries increased numbers of mobile elements across diverse families as compared to P. sojae and P. ramorum, with ∼5 times as many LTR retrotransposons and ∼10 times as many helitrons (Supplementary Fig. 7).
Consistent with a model of repeat-driven expansion of the P. infestans genome, the vast majority of repeat elements in the genome are highly similar to their consensus sequences, indicating a high rate of recent transposon activity (Supplementary Fig. 8). In addition, we have observed and experimentally confirmed examples of recently active elements (Supplementary Figs 9–11).
Phytophthora species, like many pathogens, secrete effector proteins that alter host physiology and facilitate colonization. The genome of P. infestans revealed large complex families of effector genes encoding secreted proteins that are implicated in pathogenesis10. These fall into two broad categories: apoplastic effectors that accumulate in the plant intercellular space (apoplast) and cytoplasmic effectors that are translocated directly into the plant cell by a specialized infection structure called the haustorium11. Apoplastic effectors include secreted hydrolytic enzymes such as proteases, lipases and glycosylases that probably degrade plant tissue; enzyme inhibitors to protect against host defence enzymes; and necrotizing toxins such as the Nep1-like proteins (NLPs) and PcF-like small cysteine-rich proteins (SCRs) (Supplementary Table 6).
As in the other Phytophthora species5, candidate effector genes are numerous and typically expanded compared to non-pathogenic relatives (Supplementary Table 6). Most notable among these are the RXLR and Crinkler (CRN) cytoplasmic effectors, described later.
The archetypal oomycete cytoplasmic effectors are the secreted and host-translocated RXLR proteins12. All oomycete avirulence genes (encoding products recognized by plant hosts and resulting in host immunity) discovered so far encode RXLR effectors, modular secreted proteins containing the amino-terminal motif Arg-X-Leu-Arg (in which X represents any amino acid) that defines a domain required for delivery inside plant cells11, followed by diverse, rapidly evolving carboxy-terminal effector domains13,14. Several of these C termini have been shown to exhibit virulence activities as host cell death suppressors15,16. We exploited the known motifs and other conserved sequence features to predict 563 RXLR genes in the P. infestans genome (Supplementary Tables 6, 7 and Supplementary Information). RXLR genes are notably expanded in P. infestans, with ∼60% more predicted than in P. sojae and P. ramorum (Supplementary Tables 6 and 7). We observed that 70 of these are rapidly diversifying (Supplementary Table 8). Approximately half of P. infestans RXLRs are lineage-specific, largely accounting for the expanded repertoire (Supplementary Figs 12 and 13). In contrast to the core proteome, RXLR genes show evidence of high rates of turnover with only 16 of the 563 genes with 1:1:1 orthology relationships (Supplementary Table 2) and many (88) putative RXLR pseudogenes (Supplementary Table 9). This high turnover in Phytophthora is probably driven by arms-race co-evolution with host plants5,13,14,17.
RXLR effectors show extensive sequence diversity. Markov clustering (TribeMCL18) yields one large family (P. infestans: 85, P. ramorum: 75, P. sojae: 53) and 150 smaller families (Supplementary Fig. 14). The largest family shares a repetitive C-terminal domain structure (Supplementary Figs 15 and 16). Most families have distinct sequence homologies (Supplementary Fig. 14) and patterns of shared domains (Supplementary Fig. 17) with greater diversity than expected if all RXLR effectors were monophyletic.
In contrast to the core proteome, RXLR effector genes typically occupy a genomic environment that is gene sparse and repeat-rich (Fig. 2g and Supplementary Figs 18 and 19). The mobile elements contributing to the dynamic nature of these repetitive regions may enable recombination events resulting in the higher rates of gene gain and gene loss observed for these effectors.
CRN cytoplasmic effectors were originally identified from P. infestans transcripts encoding putative secreted peptides that elicit necrosis in planta, a characteristic of plant innate immunity19. Since their discovery, little had been learned about the CRN effector family. Analysis of the P. infestans genome sequence revealed an enormous family of 196 CRN genes of unexpected complexity and diversity (Supplementary Table 10), that is heavily expanded in P. infestans relative to P. sojae (100 CRNs) and P. ramorum (19 CRNs) (Supplementary Table 6). Like RXLRs, CRNs are modular proteins. CRNs are defined by a highly conserved N-terminal ∼50-amino-acid LFLAK domain (Supplementary Fig. 20) and an adjacent diversified DWL domain (Fig. 3a, b). Most (60%) possess a predicted signal peptide. Those lacking predicted signal peptides are typically found in CRN families containing members with secretion signals (Supplementary Table 10). CRN C-terminal regions exhibit a wide variety of domain structures, with 36 conserved domains and a further eight unique C termini identified among the 315 Phytophthora CRN proteins (Supplementary Table 11). We observed evidence of recombination between different clades as a mechanism driving CRN diversity (Supplementary Figs 21–23).
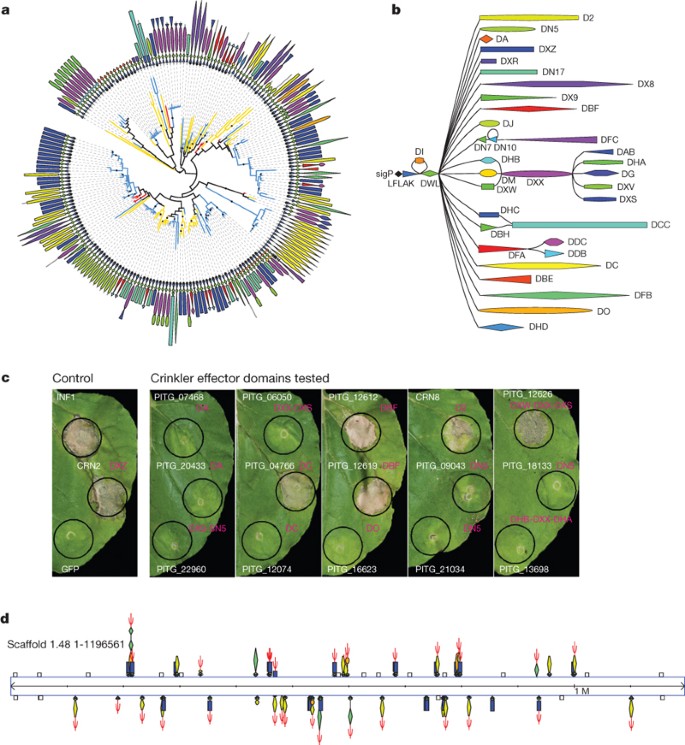
Figure 3: Diverse Crinkler (CRN) families exhibit necrosis phenotypes in planta.
a, CRN family phylogeny on the basis of the conserved N-terminal sequence, computed using PhyML with default parameters and 100 bootstrap replicates. CRN C-terminal domain structures are shown along the circumference. Branches are coloured according to organism: P. infestans in blue, P. sojae in yellow, and P. ramorum in red. Internal nodes with ≥80% bootstrap support are marked with a black dot. b, Graphical representation of the CRN family domain architecture, exhibiting a conserved N-terminal region followed by diverse C-terminal domains. c, Phenotypes observed on Nicotiana benthamiana leaves upon in planta overexpression of CRN effectors. C-terminal effector domains of CRNs were tested for cell death phenotypes on N. benthamiana leaves by _Agrobacterium tumefaciens_-mediated transient expression of CRNs, inf1 (positive control), crn2 (positive control), and green fluorescent protein (GFP) (negative control). The domains DC, DBF, D2 and DXW-DXX-DXS, like the DXZ domain of crn2, were found to induce necrosis. Cell death phenotypes were visible at 4 days post infiltration. Photos were taken 7 days after infiltration. d, CRNs with necrosis domains D2 and DXZ along with pseudogene copies are found co-clustered across P. infestans scaffold 1.48 (∼1.2 Mb). Genes and domain structures are illustrated according to the top and bottom strands of the genomic scaffold. Pseudogenes are indicated by Ψ; non-CRN genes are shown as unfilled boxes.
We explored the ability of diverse CRNs to perturb host cellular processes. In assays for necrosis in planta (Supplementary Information), deletion mutants of the previously described CRN2 secreted protein19 defined a C-terminal 234 amino-acid region (positions 173–407, domain DXZ) that is sufficient to induce cell death when expressed inside plant cells (Supplementary Fig. 24). Assays with representative P. infestans CRN genes identified four other distinct C termini that also trigger cell death inside plant cells (Fig. 3c). These include the newly defined DC domain (P. infestans: 18 genes and 49 pseudogenes (ψ)) and the D2 (14 and 43ψ) and DBF (2 and 1ψ) domains, which have similarity to protein kinases (Supplementary Table 11). These results indicate that the CRN protein domains expressed in planta are retained (lacking signal peptides and hence not secreted) by the plant cell and stimulate cell death by an intracellular mechanism, supporting the view that CRNs, like RXLRs, are cytoplasmic effectors. We propose that the conserved CRN N-terminal LFLAK domain may function similarly to the RXLR motif for delivery of CRN effectors into plant cells, and experiments to test this hypothesis are under way.
A further 255 CRN genes are fragmented or otherwise disrupted and presumably non-functional (Supplementary Table 10). CRN genes and pseudogenes are aggregated in large clusters at several genomic loci, typically clustered by domain type (Supplementary Fig. 25). One extraordinary example is scaffold 1.48 (∼1.2 Mb), containing 21 CRN genes and 31 CRN pseudogenes of the DXZ and D2 necrosis inducing domain-types (Fig. 3d). Many of the pseudogenes show only a few base changes, indicating recent conversion to pseudogenes. This high degree of expansion and pseudogene formation suggests that, like RXLR effector genes, CRN genes have undergone relatively rapid birth and death evolution.
Both CRN and RXLR genes typically occur in repeat-rich, gene-sparse regions of the genome, where conserved gene order with P. sojae and P. ramorum is either absent or disrupted (Fig. 2g, h and Supplementary Fig. 19). Expansion of large RXLR and CRN effector gene families seems to have been driven by non-allelic homologous recombination and tandem gene duplication. Although the genome is heavily populated by mobile elements, no direct evidence of transposition of effector genes was observed. Instead, the repeat-rich regions of effector clusters probably facilitate non-allelic-homologous-recombination-based expansion. In one intriguing case, nearly identical tandem arrays of CRNs are present on scaffold 1.6 in a perfect head-to-tail arrangement that is similar to that observed for some helitrons (Supplementary Fig. 26). This region of the genome is heavily enriched for helitron elements, implicating helitron-based rolling circle replication as a possible mechanism for establishing this CRN cluster.
To explore transcriptional responses to plant infection, we constructed a NimbleGen microarray based on the genome annotation. P. infestans gene expression during potato infection was monitored using samples from infected potato at 2–5 days post-inoculation (d.p.i.). In all, 494 genes were induced at least twofold during infection relative to mycelial growth. Days 2–4 of infection correlate with formation of infectious structures called haustoria. Mycelial necrotrophic growth on dead plant material occurs later at 5 d.p.i., and shows a similar expression profile to mycelial growth in plant extract media (Supplementary Fig. 27a and Supplementary Table 12). Seventy-nine RXLR genes exhibited this pattern of expression, including previously studied avirulence genes Avr3a (ref. 20), Avr4 (ref. 21), and Avr-blb1 (also known as ipiO) (ref. 22) (Supplementary Fig. 27b). Apoplastic effector genes, including protease inhibitors, cysteine-rich secreted proteins, and NPP1-family members, were among the most highly upregulated genes during infection of potato. Few CRNs were induced during infection; however most CRNs were very highly expressed, with ∼50% of CRNs within the top 10% of gene expression intensities (Supplementary Fig. 28). Several genes encoding metabolic enzymes were upregulated in planta (Supplementary Table 12), suggesting considerable metabolic adaptation of the pathogen to the host environment23. A related pattern of downregulation mirrors the induction of effectors, involving ∼115 genes (Supplementary Table 12). Among those repressed were elicitin-like genes and pseudogenes, suggesting that reduced expression during infection or mutation to pseudogene could contribute to evading activation of host innate immunity24.
P. infestans remains a critical threat to world food security, and the genome sequence is a key tool to understanding its pathogenic success. The sequence of the P. infestans genome showed an extremely high repeat content (∼74%) and unusual discontinuous distribution of gene density that correlate intriguingly with its biology. Gene-dense regions with conserved gene order across Phytophthora species are interrupted by repeat-rich expanded regions that are sparsely populated with genes, many of which are fast-evolving pathogenicity effectors such as the RXLR and CRN families. The localization of the effectors to dynamic regions of the genome probably both enables the rapid evolutionary changes and accounts for the considerable expansion in CRN and RXLR effector genes observed in P. infestans. This expansion provides a species-specific repertoire of effector genes, the dynamic nature of which probably provides an advantage in the arms race with host species. We postulate that these dynamic regions promote the evolutionary plasticity of effector genes, generating the enhanced genetic variation required to drive the rapid evasion of plant resistance that is a hallmark of the potato late blight pathogen.
Methods Summary
Genomic sequence and gene annotations
The updated P. infestans genome sequence and annotation can be accessed through GenBank accession number AATU01000000, and are available through the Broad Institute website at http://www.broad.mit.edu/annotation/genome/phytophthora_infestans. All genome sequence reads have been deposited in the NCBI trace repository (http://www.ncbi.nlm.nih.gov/Traces/home/). Paired reads of P. infestans cDNAs are available in dbEST with accessions in the range GR284383–GR301386. The NimbleGen microarray data are available in GEO under accession number GSE14480. Full methods description and associated references are provided as Supplementary Information.
References
- Reader, J. Potato: A History of the Propitious Esculent (Yale Univ. Press, 2009)
Google Scholar - Haverkort, A. J. et al. Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Res. 51, 47–57 (2008)
Article Google Scholar - Fry, W. Phytophthora infestans: the plant (and R gene) destroyer. Mol. Plant Pathol. 9, 385–402 (2008)
Article Google Scholar - McDonald, B. A. & Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379 (2002)
Article CAS Google Scholar - Tyler, B. M. et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266 (2006)
Article ADS CAS Google Scholar - Blair, J. E., Coffey, M. D., Park, S. Y., Geiser, D. M. & Kang, S. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet. Biol. 45, 266–277 (2008)
Article CAS Google Scholar - Haberer, G. et al. Structure and architecture of the maize genome. Plant Physiol. 139, 1612–1624 (2005)
Article CAS Google Scholar - Ma, J. & Bennetzen, J. L. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl Acad. Sci. USA 101, 12404–12410 (2004)
Article ADS CAS Google Scholar - Yuan, Y., SanMiguel, P. J. & Bennetzen, J. L. Methylation-spanning linker libraries link gene-rich regions and identify epigenetic boundaries in Zea mays . Genome Res. 12, 1345–1349 (2002)
Article CAS Google Scholar - Kamoun, S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60 (2006)
Article CAS Google Scholar - Whisson, S. C. et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450, 115–118 (2007)
Article ADS CAS Google Scholar - Morgan, W. & Kamoun, S. RXLR effectors of plant pathogenic oomycetes. Curr. Opin. Microbiol. 10, 332–338 (2007)
Article CAS Google Scholar - Jiang, R. H., Tripathy, S., Govers, F. & Tyler, B. M. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl Acad. Sci. USA 105, 4874–4879 (2008)
Article ADS CAS Google Scholar - Win, J. et al. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 19, 2349–2369 (2007)
Article CAS Google Scholar - Bos, J. I. et al. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana . Plant J. 48, 165–176 (2006)
Article CAS Google Scholar - Dou, D. et al. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20, 1118–1133 (2008)
Article CAS Google Scholar - Qutob, D. et al. Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a . PLoS One 4, e5066 (2009)
Article ADS Google Scholar - Enright, A. J., Kunin, V. & Ouzounis, C. A. Protein families and TRIBES in genome sequence space. Nucleic Acids Res. 31, 4632–4638 (2003)
Article CAS Google Scholar - Torto, T. A. et al. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res. 13, 1675–1685 (2003)
Article CAS Google Scholar - Armstrong, M. R. et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl Acad. Sci. USA 102, 7766–7771 (2005)
Article ADS CAS Google Scholar - van Poppel, P. M. et al. The Phytophthora infestans avirulence gene Avr4 encodes an RXLR-dEER effector. Mol. Plant Microbe Interact. 21, 1460–1470 (2008)
Article CAS Google Scholar - Vleeshouwers, V. G. et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One 3, e2875 (2008)
Article ADS Google Scholar - Grenville-Briggs, L. J. et al. Elevated amino acid biosynthesis in Phytophthora infestans during appressorium formation and potato infection. Fungal Genet. Biol. 42, 244–256 (2005)
Article CAS Google Scholar - Kamoun, S. et al. A gene encoding a protein elicitor of Phytophthora infestans is down-regulated during infection of potato. Mol. Plant Microbe Interact. 10, 13–20 (1997)
Article CAS Google Scholar
Acknowledgements
We thank L. Gaffney for help with figures and tables, E. Blanco and R. Guigo for training the GeneID gene prediction software, J. Crabtree for providing a Sybil (http://sybil.sf.net) software component used to render genome comparison illustrations, the Broad Institute Genome Sequencing Platform for sequence data generation, and C. Cuomo and D. Neafsey for comments on the manuscript. The project was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant numbers 2004-35600-15024 and 2006-35600-16623, and the National Science Foundation grants EF-0333274 and EF-0523670, and the Gatsby Charitable Foundation.
Author Contributions B.J.H., S.K., M.C.Z. and C.N. coordinated genome annotation, data analyses and manuscript preparation. B.J.H. and S.K. made equivalent contributions and should be considered joint first authors (listed by alphabetical order). R.H.Y.J., R.E.H., L.M.C., M.G., C.D.K., S.R., T.T.-A., T.O.B. and K.O. made major contributions to genome sequencing, assembly, analyses and production of complementary data and resources. All other authors are members of the genome sequencing consortium and contributed annotation, analyses or data throughout the project.
Author information
Author notes
- Chinnappa D. Kodira, Trudy Torto-Alalibo & Keith O’Neill
Present address: Present addresses: 454 Life Sciences, Branford, Connecticut 06405, USA (C.D.K.); Virginia Bioinformatics Institute, Virginia Polytechnic and State University, Blacksburg, Virginia 24061, USA (T.T.-A.); Biomedical Diagnostics Institute, Dublin City University, Dublin 9, Ireland (K.O.)., - Brian J. Haas and Sophien Kamoun: These authors contributed equally to this work.
Authors and Affiliations
- Broad Institute of MIT and Harvard, Cambridge, Massachusetts 02141, USA ,
Brian J. Haas, Michael C. Zody, Rays H. Y. Jiang, Robert E. Handsaker, Manfred Grabherr, Chinnappa D. Kodira, Lucia Alvarado, Sante Gnerre, Elinor K. Karlsson, LiJun Ma, Keith O’Neill, Ted Sharpe, Sean Sykes & Chad Nusbaum - The Sainsbury Laboratory, Norwich NR4 7UH, UK
Sophien Kamoun, Liliana M. Cano, Sylvain Raffaele, Tolga O. Bozkurt, Alexandra M. E. Jones, Jonathan D. G. Jones, Daniel MacLean, Sebastian Schornack, Ben Schwessinger, Cristina Silvar, David J. Studholme, Marco Thines & Joe Win - Department of Plant Pathology, The Ohio State University, Ohio Agricultural Research and Development Center, Wooster, Ohio 44691, USA,
Sophien Kamoun, Trudy Torto-Alalibo, Jorunn I. B. Bos, Cahid Cakir, Edgar Huitema, Zhenyu Liu, Jing Song & Carolyn Young - Department of Medical Biochemistry and Microbiology, Uppsala University, Box 597, Uppsala SE-751 24, Sweden,
Michael C. Zody - Laboratory of Phytopathology, Wageningen University, 1-6708 PB, Wageningen, The Netherlands ,
Rays H. Y. Jiang, Harold J. G. Meijer, Peter J. I. van de Vondervoort, Rob Weide & Francine Govers - Department of Plant Pathology and Microbiology, University of California, Riverside, California 92521, USA,
Audrey M. V. Ah-Fong, Sourav Roy & Howard S. Judelson - University of Aberdeen, Aberdeen Oomycete Laboratory, College of Life Sciences and Medicine, Institute of Medical Sciences, Foresterhill, Aberdeen AB25 2ZD, UK ,
Vicky L. Anderson, Laura J. Grenville-Briggs, Neil R. Horner, Ulrike D. Schumann, Stephan Wawra & Pieter van West - Plant Pathology Programme, Scottish Crop Research Institute, Invergowrie, Dundee DD2 5DA, UK ,
Miles R. Armstrong, Anna Avrova, Petra C. Boevink, Eleanor M. Gilroy, Leighton Pritchard & Stephen C. Whisson - University of Warwick, Wellesbourne, Warwick CV35 9EF, UK ,
Laura Baxter & Jim Beynon - Horticultural Crops Research Laboratory, USDA Agricultural Research Service, Corvallis, Oregon 97330, USA ,
Stephanie R. Bollmann & Niklaus J. Grünwald - Royal Institute of Technology (KTH), School of Biotechnology, AlbaNova University Centre, Stockholm SE-10691, Sweden
Vincent Bulone & Johanna Fugelstad - Department of Plant Pathology and Plant-Microbe Biology, Cornell University, Ithaca, New York 14853, USA,
Guohong Cai & William Fry - Center for Genome Research and Biocomputing and Department of Botany and Plant Pathology, Oregon State University, Corvallis, Oregon 97331, USA,
James C. Carrington & Noah Fahlgren - Biology Department, Lafayette College, Easton, Pennsylvania 18042, USA,
Megan Chawner, John Griffith, Karolyn Horn, Jessica McWalters, Manuel Ospina-Giraldo & Lauren Seyer - Plant Molecular Sciences Faculty of Biomedical and Life Sciences, Bower Building, University of Glasgow, Glasgow G12 8QQ, UK
Lucio Conti, Richard Ewan & Ari Sadanandom - USDA-ARS, Dale Bumpers National Rice Research Center, Stuttgart, Arkansas 72160, USA ,
Stefano Costanzo - Department of Molecular Biology, Massachusetts General Hospital, Boston, Massachsetts 02114, USA,
Michael A. Fischbach - Delaware Biotechnology Institute, University of Delaware, Newark, Delaware 19711, USA ,
Pamela J. Green, Dong-Hoon Jeong & Blake C. Meyers - Department of Plant Pathology, North Carolina State University, Raleigh, North Carolina 27695, USA,
Chia-Hui Hu & Jean Ristaino - USDA-ARS, Beltsville, Maryland 20705, USA ,
Richard W. Jones - Department of Plant and Soil Sciences, University of Delaware, Newark, Delaware 19716, USA,
Sridhara G. Kunjeti - Department of Entomology and Plant Pathology, University of Tennessee, Knoxville, Tennessee 37996, USA,
Kurt Lamour - Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, Maryland 21201, USA ,
Marcus C. Chibucos - Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, Tennessee 37232, USA,
Hayes McDonald - Department of Biology, The College of Wooster, Wooster, Ohio 44691, USA,
William Morgan - Department of Biological Sciences, Bowling Green State University, Bowling Green, Ohio 43403, USA,
Paul F. Morris & Vipaporn Phuntumart - University of Aberdeen, School of Medical Sciences, College of Life Sciences and Medicine, Institute of Medical Sciences, Foresterhill, Aberdeen AB25 2ZD, UK ,
Carol A. Munro - Mycology and Phytopathology Laboratory, Los Andes University, Bogotá, Colombia
Andrés Pinzón & Silvia Restrepo - Institute of Genetics and Molecular Medicine, University of Edinburgh, Cancer Research Centre, Western General Hospital, Edinburgh EH4 2XU, UK
Bernard Ramsahoye - J. Craig Venter Institute, Rockville, Maryland 20850, USA ,
Qinghu Ren - Department of Plant Sciences, Tel Aviv University, Tel Aviv 69978, Israel
Alon Savidor - Department of Chemistry, Laboratory of Genetics, Laboratory for Molecular and Computational Genomics, University of Wisconsin Biotechnology Center, University of Wisconsin-Madison, Madison Wisconsin 53706, USA
David C. Schwartz & Shiguo Zhou - University of Hohenheim, Institute of Botany 210, D-70593 Stuttgart, Germany
Marco Thines - Division of Plant Science, College of Life Sciences, University of Dundee (at SCRI), Invergowrie, Dundee DD2 5DA, UK,
Paul R. J. Birch
Authors
- Brian J. Haas
- Sophien Kamoun
- Michael C. Zody
- Rays H. Y. Jiang
- Robert E. Handsaker
- Liliana M. Cano
- Manfred Grabherr
- Chinnappa D. Kodira
- Sylvain Raffaele
- Trudy Torto-Alalibo
- Tolga O. Bozkurt
- Audrey M. V. Ah-Fong
- Lucia Alvarado
- Vicky L. Anderson
- Miles R. Armstrong
- Anna Avrova
- Laura Baxter
- Jim Beynon
- Petra C. Boevink
- Stephanie R. Bollmann
- Jorunn I. B. Bos
- Vincent Bulone
- Guohong Cai
- Cahid Cakir
- James C. Carrington
- Megan Chawner
- Lucio Conti
- Stefano Costanzo
- Richard Ewan
- Noah Fahlgren
- Michael A. Fischbach
- Johanna Fugelstad
- Eleanor M. Gilroy
- Sante Gnerre
- Pamela J. Green
- Laura J. Grenville-Briggs
- John Griffith
- Niklaus J. Grünwald
- Karolyn Horn
- Neil R. Horner
- Chia-Hui Hu
- Edgar Huitema
- Dong-Hoon Jeong
- Alexandra M. E. Jones
- Jonathan D. G. Jones
- Richard W. Jones
- Elinor K. Karlsson
- Sridhara G. Kunjeti
- Kurt Lamour
- Zhenyu Liu
- LiJun Ma
- Daniel MacLean
- Marcus C. Chibucos
- Hayes McDonald
- Jessica McWalters
- Harold J. G. Meijer
- William Morgan
- Paul F. Morris
- Carol A. Munro
- Keith O’Neill
- Manuel Ospina-Giraldo
- Andrés Pinzón
- Leighton Pritchard
- Bernard Ramsahoye
- Qinghu Ren
- Silvia Restrepo
- Sourav Roy
- Ari Sadanandom
- Alon Savidor
- Sebastian Schornack
- David C. Schwartz
- Ulrike D. Schumann
- Ben Schwessinger
- Lauren Seyer
- Ted Sharpe
- Cristina Silvar
- Jing Song
- David J. Studholme
- Sean Sykes
- Marco Thines
- Peter J. I. van de Vondervoort
- Vipaporn Phuntumart
- Stephan Wawra
- Rob Weide
- Joe Win
- Carolyn Young
- Shiguo Zhou
- William Fry
- Blake C. Meyers
- Pieter van West
- Jean Ristaino
- Francine Govers
- Paul R. J. Birch
- Stephen C. Whisson
- Howard S. Judelson
- Chad Nusbaum
Corresponding authors
Correspondence toSophien Kamoun or Chad Nusbaum.
Supplementary information
PowerPoint slides
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/), which permits distribution, and reproduction in any medium, provided the original author and source are credited. This license does not permit commercial exploitation, and derivative works must be licensed under the same or similar licence.
About this article
Cite this article
Haas, B., Kamoun, S., Zody, M. et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans.Nature 461, 393–398 (2009). https://doi.org/10.1038/nature08358
- Received: 23 April 2009
- Accepted: 31 July 2009
- Published: 09 September 2009
- Issue Date: 17 September 2009
- DOI: https://doi.org/10.1038/nature08358