Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice (original) (raw)
Main
Horses show considerable variation in the pattern of locomotion. The three naturally occurring gaits in all equids are, in order of increasing speed, walk, trot and canter/gallop. Some horses can use alternate gaits, typically at intermediate speed, and ‘gaitedness’ is a trait upon which many specialized breeds have been developed. Based on variation in footfall pattern, timing and cadence, these alternate gaits can be generally divided into four categories: pace, regular rhythm ambling, lateral ambling and diagonal ambling (Supplementary Notes and Supplementary Table 1). Pace is a two-beat gait in which the horse moves the two legs on the same side of the body in a synchronized, lateral movement (Fig. 1a) in contrast to the trot, where the diagonal front and hind legs move forward and backward together (Fig. 1b). Ambling gaits are four-beat gaits in which footfall pattern, foot placement and timing are often unique to specific breeds (Supplementary Notes and Supplementary Table 1). Tölt is a regular ambling gait characteristic of the Icelandic horse. Many Icelandic horses also have the ability to pace and test scores for pace show a bimodal distribution (Fig. 1c) and high heritability, in the range 0.60–0.73 (ref. 3).
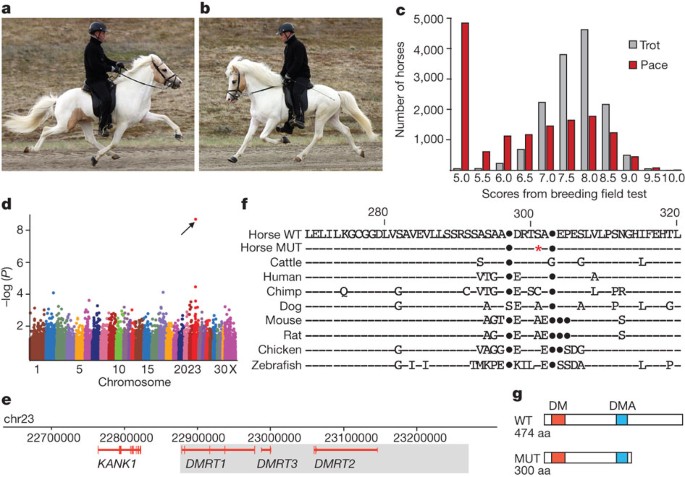
Figure 1: Identification of a DMRT3 mutation in horses.
a, A pacing Icelandic horse, fore- and hindlegs on the same side of the body are synchronized. b, A trotting Icelandic horse, the diagonal fore- and hindlegs are synchronized. c, Distribution of breeding evaluation test scores for pace and trot in Icelandic horses. Score 5.0 indicates ‘gait not shown’. d, Genome-wide association analysis revealed a highly significant association between the ability to pace and SNP BIEC2_620109 on chromosome 23 (_P_raw = 1.7 × 10−9, corrected empirical _P_-value (EMP2) = 2.0 × 10−4, genome-wide significance). e, The 684 kb genomic interval associated with the Gait locus; the minimum Gait IBD region (438 kb) is shaded. f, Partial amino acid alignment of the predicted Dmrt3 protein in wild-type (WT) and mutant (MUT) horses and in other vertebrates. Horse nonsense mutation (red asterisk), sequence identities (dashes), insertions/deletions (dots). g, Schematics of wild-type and mutant Dmrt3. DM, zinc-finger like DNA binding module; DMA, protein domain of unknown function present in DMRT proteins.
A genome-wide association analysis using 30 Icelandic horses classified as four-gaited (walk, tölt, trot and gallop) and 40 classified as five-gaited (walk, tölt, trot, gallop and pace) revealed a highly significant association between the ability to pace and a single nucleotide polymorphism (SNP; BIEC2_620109) at nucleotide position 22967656 on chromosome 23 (Fig. 1d). The two flanking markers showed only weak association to the pacing phenotype, indicating that the mutation(s) underlying the association is located within the 684-kilobases interval chr23:22628976–23315071. We resequenced selected regions of the 684-kb interval in a panel of four- and five-gaited Icelandic horses. The five-gaited horses were all homozygous for a minimal 438-kb haplotype (chr23:22877015–23315071), that was inferred to be identical-by-descent (IBD; Supplementary Table 2). This region contains only three genes encoding different isoforms of the doublesex and mab-3 related transcription factors, DMRT1-3 (Fig. 1e). The DMRT family of transcription factors carry a DM (dsx and mab-3) DNA-binding domain, conferring sequence-specific DNA binding distinct from a classical zinc-finger4.
We performed whole-genome resequencing of one four-gaited and one five-gaited Icelandic horse, homozygous for opposite alleles at the SNP associated with the ability to pace. Average sequence coverage of 30× was obtained and polymorphisms identified in the critical 438-kb interval were compiled (Supplementary Table 3). Homozygosity mapping using the sequenced five-gaited horse confirmed an IBD region of about 438 kb. In this interval, we identified 65 sequence differences (60 SNPs and five small insertions/deletions) unique to the five-gaited horse when comparing data for the two horses and the reference genome (Supplementary Table 4); no structural rearrangements were detected. We found five intronic or intragenic SNPs at sites showing some degree of evolutionary conservation, and a single base change at nucleotide position chr23:22999655 causing a premature stop at codon 301 in DMRT3 (DMRT3_Ser301STOP; Fig. 1f). The allele is expected to encode a truncated protein lacking 174 amino acid residues of the full-length protein (Fig. 1g), of which 161 (92.5%) are identical between human and horse Dmrt3. DMRT3_Ser301STOP was evaluated as the candidate causative mutation.
We genotyped 352 additional Icelandic horses and found that all but one of the five-gaited horses were homozygous A/A for the DMRT3 nonsense mutation (Table 1); further investigation of competition records revealed that this single discordant horse was most likely phenotypically misclassified. In contrast, only 31% of the four-gaited horses were homozygous A/A (P = 2.4 × 10−14). Thus, homozygosity for the DMRT3 nonsense mutation is required for the ability to pace in this breed. The observation that a considerable number of homozygous mutant horses are considered four-gaited may reflect phenotype misclassifications, but more likely incomplete penetrance due to other genetic factors, maturity and environmental effects, in particular training.
Table 1 Allele frequency of the DMRT3 nonsense mutation among horse populations
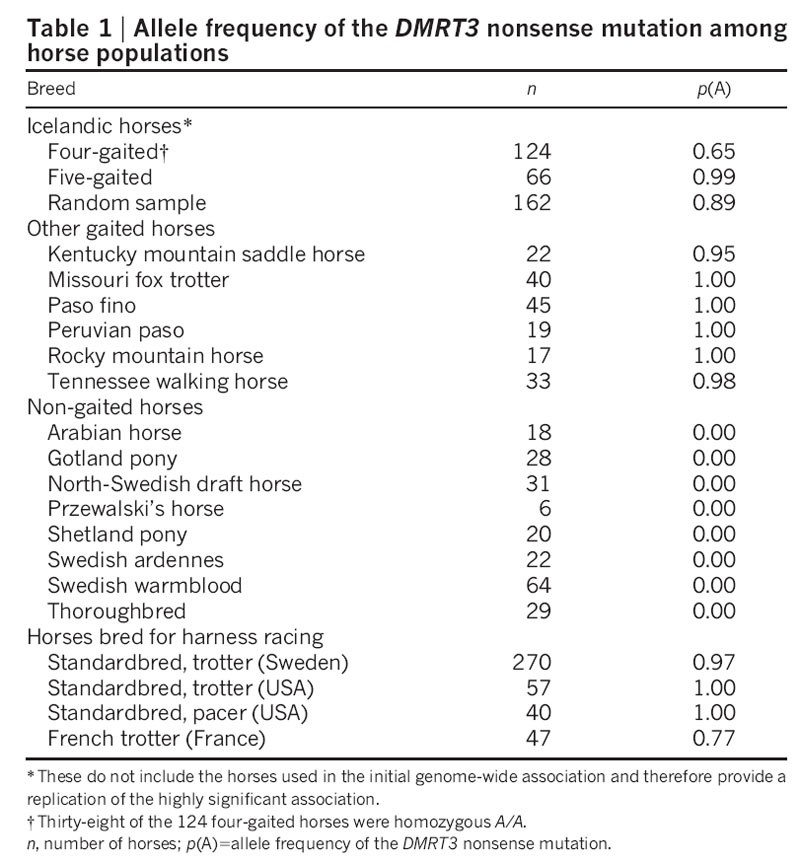
The DMRT3 genotype distribution across breeds was markedly dichotomous, with the mutation occurring at high frequency in all gaited breeds, whereas all tested non-gaited horses were homozygous wild type (Table 1), with the exception of horses used for harness racing (see below). Nearly all individuals from other gaited breeds were homozygous mutant, regardless of whether their four-beat alternate gait is characterized by lateral or diagonal couplets (Supplementary Table 1). Thus, the DMRT3 mutation is permissive for the ability to perform alternate gaits, which can be either pace or four-beat ambling gaits. Although this mutation must be advantageous for gaited horses, it may be disadvantageous for others. In fact, Icelandic horse homozygous mutants had inferior scores for gallop and trot (Supplementary Table 5). Thus, there may be selection against the mutation in non-gaited horses bred for dressage, show jumping or high-speed gallop.
We found a high frequency of the DMRT3 mutation in horses bred for harness racing (Table 1). These horses have the ability to trot or pace at high speed without breaking into a gallop, the natural gait at high speed for horses. The American Standardbred was established in the 19th century and bred for harness racing. Competitions are held separately in trot or pace and assortative mating based on preferred gait has subdivided the breed into two populations, pacers and trotters. In contrast to the pattern in Icelandic horses, where homozygosity for DMRT3_Ser301STOP was associated with the ability to pace, both Standardbred pacers and trotters are homozygous for the mutation. Thus, the mutation may promote the ability to trot or pace at high speed and genetic modifiers determine the gait to which the horse is best suited.
The Swedish Standardbred is largely developed from the American Standardbred but is not completely fixed for the DMRT3 mutation, probably owing to the import of French trotters, a breed with a fairly high frequency of the wild-type allele (Table 1). The segregation of the two alleles in the Swedish Standardbred provided an opportunity to examine the effect of the mutation on racing performance. The DMRT3 mutation was associated with superior breeding values (BV) for racing performance (BVCA = 95.7 ± 1.7, n = 17; BVAA = 109.0 ± 0.8, n = 206; P < 0.0001) and increased earned prize money (_X_CA = 48,000 ± US$35,000, n = 17; _X_AA = 161,000 ± US$24,000, n = 206; _P_one-sided = 0.007). We also genotyped 61 horses from one racing camp in a blind test; two of these had major difficulties in sustaining trot at high speed (Supplementary Fig. 1a, b and Supplementary Movie 1) and were heterozygous C/A, whereas all others were homozygous A/A (P = 0.0005).
Whereas the horse discovery demonstrates that DMRT3 has an effect on gait coordination, studies of its possible role in locomotor circuitry are more tractable in mice. We used _Dmrt3_-null mice5 (Supplementary Fig. 2) and evaluated locomotion performances. Motor coordination and balance were largely normal in Dmrt3 −/− mice (Supplementary Fig. 3a–c). In water, Dmrt3 −/− mice spent less time swimming and showed frequent twitching limb movements rarely observed in controls (Fig. 2a and Supplementary Movie 2). Next, mice were placed on a TreadScan apparatus, which performs an automated and unbiased analysis to collect multiple gait parameters (Supplementary Table 6). Dmrt3 −/− mice, but not control mice (wild-type littermates), had major difficulties running at higher velocities (Fig. 2b and Supplementary Movie 3). Gait analysis (Fig. 2c) revealed significantly increased stride length in all limbs of Dmrt3 −/− mice (Fig. 2d). Swing times (flexion) were increased in all limbs, whereas stance time (extension) was increased in forelimbs (Fig. 2e, f). Moreover, propulsion time increased in all limbs, whereas brake time was decreased in hindlimbs, indicating that Dmrt3 −/− mice may emphasize extension movements, resulting in a longer stride (Supplementary Table 6). Heterozygotes did not differ significantly from controls.
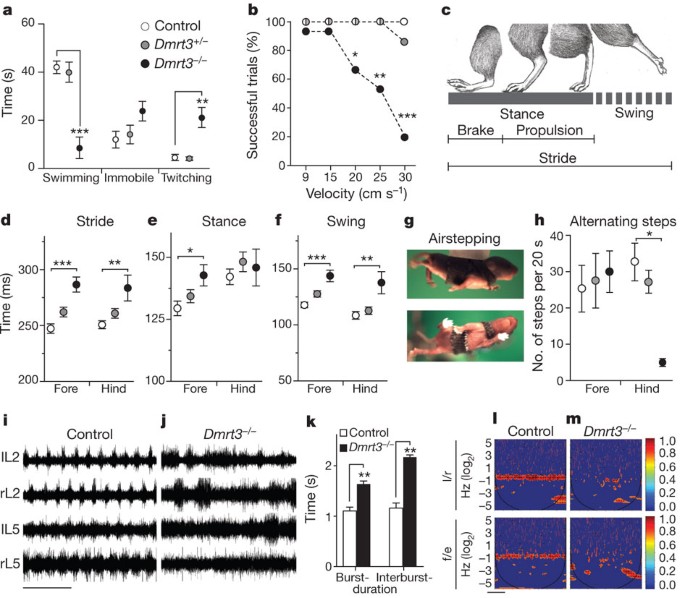
Figure 2: Characterization of motor coordination in mice lacking Dmrt3.
a, _Dmrt3_−/− mice showed decreased swimming duration (P < 0.0001), and an increase in twitching movements compared to control and _Dmrt3_+/− (_P_ = 0.0002); _n_ = 5 control and _Dmrt3_−/−, _n_ = 7 _Dmrt3_+/−. Time spent immobile was similar between genotypes (_P_ = 0.13). b**, Mice were tested for their ability to run (>5 step cycles) at different treadmill speeds (9, 15, 20, 25, 30 cm s−1) (n = 15 trials per genotype, five animals). _Dmrt3_−/− mice had difficulties at 20 cm s−1 (P = 0.04) with markedly reduced number of successful trials at 25 cm s−1 (P = 0.006) and 30 cm s−1 (P < 0.0001) compared to controls. c, Schematic drawing of the gait parameters analysed in treadmill locomotion. d–f**, At 20 cm s−1 _Dmrt3_−/− showed increased stride (d, forelimb: P < 0.0001, hindlimb: P = 0.006), hind limb stance (e, P = 0.02) and swing (f, forelimb: P < 0.0001, hindlimb: P = 0.001) time duration compared to control (n = 11–12 trials per genotype, five animals). g, Images of a P4 mouse airstepping. h, _Dmrt3_−/− mice showed a decreased number of alternating hindlimb steps during airstepping (P = 0.02 compared to controls; n = 4 per genotype, except controls n = 3). Wild-type littermate controls (white), Dmrt3+/− (grey), _Dmrt3_−/− (black). Mean ± s.e.m. i–m, Fictive locomotion was recorded from ventral left (l) and right (r) lumbar (L) root 2 and 5 from neonatal spinal cords. i, j, Representative traces from control (i) and _Dmrt3_−/− (j) spinal cords. Time scale 10 s. k, _Dmrt3_−/− mice displayed increased burst and interburst durations compared to control animals, analysis made on L2s (n = 6 per genotype, P = 0.02, burst; P = 0.001, interburst). l, m, Coherence power spectra analysis of left/right (l/r) and flexor/extensor (f/e) recordings (colour-graded scale; n = 3–5 per genotype). Time scale 100 s.
Development of weight-supported walking was similar in wild-type and Dmrt3 −/− mice (Supplementary Fig. 3d, e), whereas limb coordination in neonatal mice, scored during air-stepping, was markedly different (Fig. 2g). At postnatal (P) day 4, we observed similar numbers of alternating step movements in all wild-type limbs as well as in Dmrt3 −/− mice forelimbs. However, alternating hindlimb movements were almost absent in Dmrt3 −/− mice (Fig. 2h), accompanied by increased uncoordinated step movements (Supplementary Fig. 3f). This early effect was emphasized by a similar phenotype at P1 (Supplementary Fig. 3g–i). Next, we analysed central pattern generator output in the isolated neonatal spinal cord using drug-induced fictive locomotion. Cords from wild-type mice generated a stable rhythm, whereas cords from Dmrt3 −/− mice had uncoordinated and irregular firing rhythms as well as increased burst and interburst durations (Fig. 2i–k). Moreover, the coefficient of variation, a normalized measure of variability, increased two- to threefold (Supplementary Fig. 4). We analysed the rhythm relationship between the left-right (l/r) and flexion-extension (f/e) outputs using a continuous wavelet transform that measures the coherence between the spinal ventral roots with high resolution6,7. Wild-type and heterozygous mice had coherence frequencies around 0.4 Hz with clear coordinated left-right and flexion-extension alternation (coherence in wild-type l/r 95%, f/e 92%). In contrast, Dmrt3 −/− mice showed more variable and lower frequency values (≈0.1 Hz), and bursts were non-coherent between left-right and flexion-extension (Fig. 2l, m). The coherence values were lower (l/r 23%, f/e 25%) and significantly reduced compared to wild-types (P = 0.01, l/r; P = 0.01, f/e). The remaining coherent activity showed no clear direction towards synchrony or alternation (Supplementary Fig. 4). Excitation-inhibition balance can be influenced by the glycine re-uptake inhibitor sarcosine8; however, the collapsed coordination observed here was unaffected by such treatment. Moreover, strychnine-induced synchronous bursting was similar between genotypes, indicating that excitatory cross coupling was present in Dmrt3 −/− mice (Supplementary Fig. 4).
Post-natally, Dmrt3 messenger RNA was expressed in the ventral spinal cord at all levels, whereas prenatal expression was evident in more dorsally located cells, indicative of dorsoventral migration (Fig. 3a, b and Supplementary Fig. 5). Already at embryonic day (E) 12.5, Dmrt3 cells were found in the medioventral domain, a pattern that persisted in adults. To pinpoint the origin of Dmrt3 cells, we used markers for dorsal and ventral progenitors giving rise to different subclasses of interneurons9 (Fig. 3c). Immunostainings at E11.5 for Dmrt3 and Pax7, a marker for the dorsal progenitor domain10, demonstrated that Dmrt3 immunopositive (+) cells are generated near the ventral Pax7+ domain border (Fig. 3d). Moreover, Dmrt3+ cells were found within the Pax2 domain marking dI4, dI5 and V0d progenitors11, while they were negative for Evx1, a post-mitotic marker for V0v interneurons12. Dmrt3 expression did not coincide with the dI5 marker Lmx1b13 whereas labelling with the dI4-6 postmitotic marker Lbx1 (ref. 14) indicated that the Dmrt3+ population arise from the ventral-most Lbx1 domain (Fig. 3d). These data indicate that Dmrt3 marks a dI6 interneuron subpopulation, further confirmed by in situ hybridization analysis and additional markers (Supplementary Fig. 5). At E14.5, we found a partial overlap with Wt1 (Fig. 3e), proposed to label the dI6 population15. Dmrt3+ interneurons in P11 spinal sections were positive for Viaat (also known as Slc32a1), marking inhibitory neurons, but negative for Vglut2 (also known as Slc17a6), marking the majority of spinal excitatory neurons16 (Fig. 3f). Fluorescein-dextran retrograde tracings in E15 spinal cords17 revealed Dmrt3+ interneurons that extended projections ipsilaterally (22%) and contralaterally (39%). Contralateral fibres were not observed at E12.5, indicating that midline crossing occurs between E12.5 and E15.5 (Supplementary Fig. 6). Trans-synaptic pseudorabies-virus tracing in hindlimb muscles was used to examine whether Dmrt3 interneurons contact motor neurons (Fig. 3g). Forty hours post-infection, we found virus+/Dmrt3+ cells both ipsi- and contralateral to the injected muscle, indicating direct connections to motor neurons18 and corroborating the presence of commissural fibres from Dmrt3+ cells (Fig. 3h). Although their character may change in the adult mouse, our developmental analysis suggests that Dmrt3-expressing cells originate from dI6 progenitors at around E11.5, develop into inhibitory interneurons with projecting axons ipsi- and contralateral, and make synaptic connections to motor neurons.
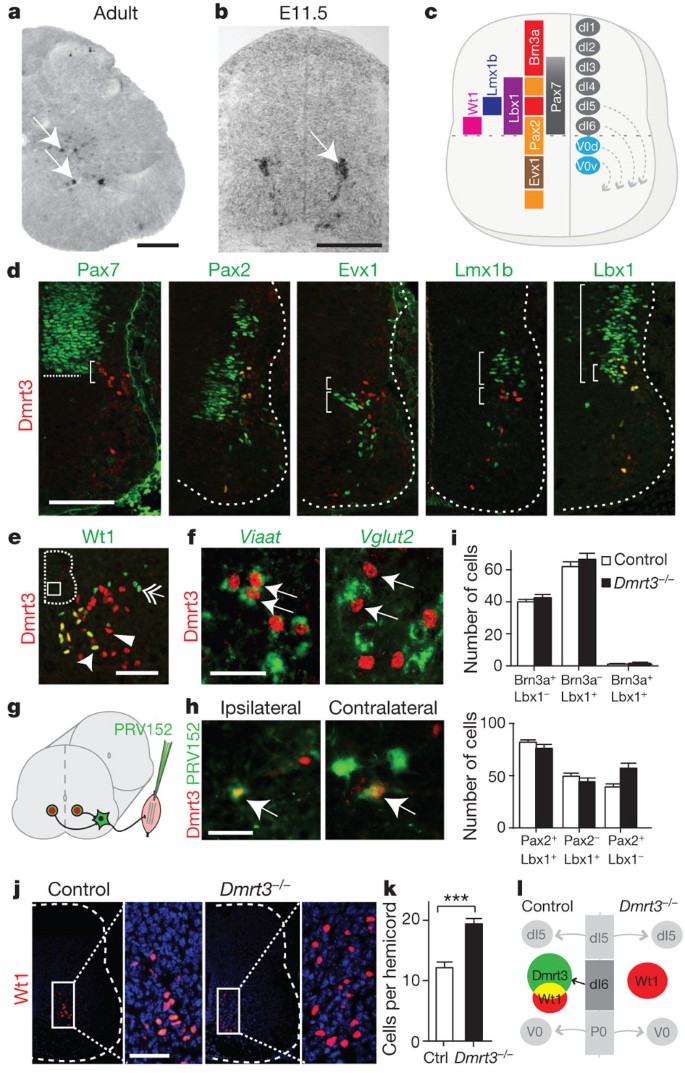
Figure 3: Characterization of Dmrt3-expressing cells in the mouse spinal cord.
a, Dmrt3 mRNA expression pattern in adult spinal cord (P60). b, Dmrt3 mRNA expression in a restricted population of neurons migrating ventrally in the developing spinal cord at E11.5. c, A schematic spinal cord cross-section showing progenitor and transcription factor domains. d, Double immunolabelling of Dmrt3 and Pax7 shows that Dmrt3+ cells originate from the ventral-most part (bracket) of the dorsal domain (border indicated by line). Dmrt3+ cells overlap with the dI4/dI6/V0d marker Pax2, but not with the V0V/V0C/V0G marker Evx1 or the dI5 marker Lmx1b (compare brackets). Dmrt3+ cells overlap with the dI4/dI5/dI6 marker Lbx1. e, Double immunolabelling with Dmrt3 (arrowhead) and Wt1 (double arrow) show a partial overlap (arrow). f, Dmrt3+ interneurons (arrows) co-labelled with Viaat mRNA (green) but not with Vglut2 mRNA (green). g, Schematic of trans-synaptic muscle tracing of Dmrt3 neurons in the spinal cord (n = 6, ipsilateral; n = 5, contralateral). h, Double immunolabelling of Dmrt3 and green fluorescent protein (PRV152) show that both ipsi- and contralateral premotor interneurons overlap with Dmrt3+ cells (arrows). i, Quantification of Brn3a/Lbx1-positive neurons (control _n_section = 16, _Dmrt3_−/− _n_section = 21) and of Lbx1/Pax2-positive neurons (control _n_section = 11, _Dmrt3_−/− _n_section = 19). j, Immunolabelling of Wt1+ cells (red) in spinal cord sections from control and _Dmrt3_−/− E15.5 embryos. k, Quantification demonstrated that loss of Dmrt3 leads to an expanded Wt1+ cell subpopulation (control n = 43, Dmrt3 +/− n = 36, Dmrt3 −/− n = 46, ***P < 0.0001). l, Schematic illustration of fate change in the Dmrt3 dI6 population of neurons. Mean ± s.e.m. Scale bars: 400 μm (a), 70 μm (b, d, j), 50 μm (e, f, h).
Because loss of Dmrt3 may affect interneuron development in mice, we next analysed the dI6 population, and the flanking dI5 and V0d populations, by Pax2, Brn3a and Lbx1 immunostainings. All three populations remained of similar sizes in wild-type and _Dmrt3_−/− mice (Fig. 3i). In contrast, we found a 58% increase in the number of Wt1+ neurons in _Dmrt3_−/− mice (Fig. 3j–l, P < 0.0001), suggesting a fate change within a specific subset of dI6 neurons. Moreover, retrograde tracing in E15 and P0 animals revealed significant decreases in commissural interneuron numbers in _Dmrt3_−/− mice compared to wild-type at E15 (P = 0.001) and P0 (P = 0.005) (Supplementary Fig. 6). Thus, loss of Dmrt3 resulted in an increased number of Wt1+ cells and fewer commissural interneurons, probably explained by an altered fate of the Dmrt3+ population. In general, loss of transcription factor expression within progenitor domains results in neuron specification defects, presumably by suppression of differentiation programs operating in adjacent domains15,19,20,21. Our results suggest an early reprogramming of spinal interneurons; however, circuit reorganization, compensation issues and the direct role of Dmrt3+ interneurons require further investigation.
As the horse DMRT3 mutation occurs in the last exon, the mRNA is not expected to be subject to nonsense-mediated RNA decay22 and the mutant allele is probably translated into a truncated form (Fig. 1g). Expression levels between mutant and wild-type homozygotes were similar and DMRT3 mRNA was found in a small population of neurons located in the ventral horn and around the central canal in both wild-type and mutant horses (Supplementary Fig. 1c–e). Furthermore, transfection experiments and an electrophoretic mobility shift assay indicated that the mutant Dmrt3 protein maintain its cellular localization and DNA-binding profile (Supplementary Fig. 1f, g). It may therefore be a dominant negative form with normal DNA binding but defective protein interactions.
The remarkable association between the DMRT3 nonsense mutation and gaitedness across horse breeds, combined with the demonstration that mouse Dmrt3 is required for normal development of a coordinated locomotor network in the spinal cord, allow us to conclude that DMRT3_Ser301STOP is a causative mutation affecting the pattern of locomotion in horses. The horse phenotype indicates that Dmrt3 neurons not only have a critical role for left/right coordination but also for coordinating the movement of the fore- and hindlegs. The mutation facilitates lateral gaits, ambling and pace, and inhibits the transition from trot or pace to gallop. Homozygosity for the mutation is required, but not sufficient for pacing, as many Standardbred trotters and some Icelandic horses that are homozygous mutant do not pace. In mice, a complete loss of Dmrt3 on one allele does not lead to any detectable phenotype, whereas in Icelandic horses, heterozygosity for the DMRT3_Ser301STOP mutation promotes tölt, supporting our hypothesis that the mutant protein in horses acts as a dominant negative form. Dmrt3 neurons are present in the horse and mouse spinal cord, and in the mouse, they develop into premotor inhibitory interneurons projecting ipsi- and contralaterally. Inhibitory commissural connections have been suggested as major constituents of left-right phasing during locomotion23,24,25, and in cat, such interneurons have been implicated in mediation of the crossed reflexes and in the selection of different motor patterns26,27. Thus, Dmrt3 neurons have a character and position in spinal cord circuitry that concurs with gait coordination.
Methods Summary
A summary of the methods can be found in the Supplementary Information and includes detailed information on study populations, genotyping methods and genome-wide association analysis, genome resequencing and calling of genetic variants, _Dmrt3_-null mice, immunohistochemistry, in situ hybridization of mouse and horse tissue, spinal cord and muscle tracing, extracellular physiology, behaviour recordings and statistical analyses, expression analysis using mouse and horse tissue, transfection experiments and electrophoretic mobility shift assays.
Accession codes
Primary accessions
Sequence Read Archive
Data deposits
The Illumina reads have been submitted to the short reads archive (http://www.ncbi.nlm.nih.gov/sra); the accession number for the study is SRP012260 and accession numbers for individual data are: four-gaited horse, SRS309533; five-gaited horse, SRS309532. Sanger sequencing data have been submitted to GenBank (accession numbers JQ922365–JQ922395).
References
- Grillner, S. Biological pattern generation: the cellular and computational logic of networks in motion. Neuron 52, 751–766 (2006)
Article CAS Google Scholar - Kullander, K. Genetics moving to neuronal networks. Trends Neurosci. 28, 239–247 (2005)
Article CAS Google Scholar - Albertsdóttir, E., Eriksson, S., Sigurdsson, A. & Arnason, T. Genetic analysis of ‘breeding field test status’ in Icelandic horses. J. Anim. Breed. Genet. 128, 124–132 (2011)
Article Google Scholar - Hong, C. S., Park, B. Y. & Saint-Jeannet, J. P. The function of Dmrt genes in vertebrate development: it is not just about sex. Dev. Biol. 310, 1–9 (2007)
Article CAS Google Scholar - Ahituv, N. et al. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 5, e234 (2007)
Article Google Scholar - Gallarda, B. W., Sharpee, T. O., Pfaff, S. L. & Alaynick, W. A. Defining rhythmic locomotor burst patterns using a continuous wavelet transform. Ann. NY Acad. Sci. 1198, 133–139 (2010)
Article ADS Google Scholar - Mor, Y. & Lev-Tov, A. Analysis of rhythmic patterns produced by spinal neural networks. J. Neurophysiol. 98, 2807–2817 (2007)
Article CAS Google Scholar - Kullander, K. et al. Role of EphA4 and EphrinB3 in local neuronal circuits that control walking. Science 299, 1889–1892 (2003)
Article ADS CAS Google Scholar - Alaynick, W. A., Jessell, T. M. & Pfaff, S. L. SnapShot: spinal cord development. Cell 146, 178e1 (2011)
Article Google Scholar - Jostes, B., Walther, C. & Gruss, P. The murine paired box gene, Pax7, is expressed specifically during the development of the nervous and muscular system. Mech. Dev. 33, 27–37 (1990)
Article CAS Google Scholar - Burrill, J. D., Moran, L., Goulding, M. D. & Saueressig, H. PAX2 is expressed in multiple spinal cord interneurons, including a population of EN1+ interneurons that require PAX6 for their development. Development 124, 4493–4503 (1997)
CAS PubMed Google Scholar - Moran-Rivard, L. et al. Evx1 is a postmitotic determinant of V0 interneuron identity in the spinal cord. Neuron 29, 385–399 (2001)
Article CAS Google Scholar - Müller, T. et al. The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron 34, 551–562 (2002)
Article Google Scholar - Gross, M. K., Dottori, M. & Goulding, M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron 34, 535–549 (2002)
Article CAS Google Scholar - Goulding, M. Circuits controlling vertebrate locomotion: moving in a new direction. Nature Rev. Neurosci. 10, 507–518 (2009)
Article CAS Google Scholar - Gezelius, H., Wallen-Mackenzie, A., Enjin, A., Lagerstrom, M. & Kullander, K. Role of glutamate in locomotor rhythm generating neuronal circuitry. J. Physiol. Paris 100, 297–303 (2006)
Article CAS Google Scholar - Rabe, N., Gezelius, H., Vallstedt, A., Memic, F. & Kullander, K. Netrin-1-dependent spinal interneuron subtypes are required for the formation of left-right alternating locomotor circuitry. J. Neurosci. 29, 15642–15649 (2009)
Article CAS Google Scholar - Jovanovic, K., Pastor, A. M. & O’Donovan, M. J. The use of PRV-Bartha to define premotor inputs to lumbar motoneurons in the neonatal spinal cord of the mouse. PLoS ONE 5, e11743 (2010)
Article ADS Google Scholar - Lanuza, G. M., Gosgnach, S., Pierani, A., Jessell, T. M. & Goulding, M. Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron 42, 375–386 (2004)
Article CAS Google Scholar - Pierani, A. et al. Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron 29, 367–384 (2001)
Article CAS Google Scholar - Vallstedt, A. et al. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron 31, 743–755 (2001)
Article CAS Google Scholar - Culbertson, M. R. & Leeds, P. F. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13, 207–214 (2003)
Article CAS Google Scholar - Cowley, K. C. & Schmidt, B. J. Effects of inhibitory amino acid antagonists on reciprocal inhibitory interactions during rhythmic motor activity in the in vitro neonatal rat spinal cord. J. Neurophysiol. 74, 1109–1117 (1995)
Article CAS Google Scholar - Grillner, S. & Wallen, P. Does the central pattern generation for locomotion in lamprey depend on glycine inhibition? Acta Physiol. Scand. 110, 103–105 (1980)
Article CAS Google Scholar - Jankowska, E. & Noga, B. R. Contralaterally projecting lamina VIII interneurones in middle lumbar segments in the cat. Brain Res. 535, 327–330 (1990)
Article CAS Google Scholar - Harrison, P. J., Jankowska, E. & Zytnicki, D. Lamina VIII interneurones interposed in crossed reflex pathways in the cat. J. Physiol. (Lond.) 371, 147–166 (1986)
Article CAS Google Scholar - Jankowska, E., Edgley, S. A., Krutki, P. & Hammar, I. Functional differentiation and organization of feline midlumbar commissural interneurones. J. Physiol. (Lond.) 565, 645–658 (2005)
Article CAS Google Scholar
Acknowledgements
Thanks to S. Mikulovic and E. Restrepo for valuable input, C. Birchmeier for Lbx1 antibody, L. Enquist and J. Martin for PRV152, S. Ewart for horse samples, and B. Ågerup for access to race horses. The work was supported by grants from the Swedish Foundation for Strategic Research, the Swedish Research Council Formas (221-2009-1631), Swedish Research Council Medicine and Health (2007-3630/4479, 2010-4394), Swedish Society for Medical Research (H.W.), National Institute of Child Health & Human Development R01HD059862 (N.A.), and the Swedish Brain Foundation. Sequencing was performed by the SNP&SEQ Technology Platform, supported by Uppsala University and Hospital, SciLife Lab – Uppsala and the Swedish Research Council (80576801 and 70374401). Computer resources were supplied by UPPMAX. K.K. is a Royal Swedish Academy of Sciences Research Fellow supported by a grant from the Knut and Alice Wallenberg Foundation.
Author information
Author notes
- Lisa S. Andersson, Martin Larhammar, Fatima Memic, Hanna Wootz, Leif Andersson and Klas Kullander: These authors contributed equally to this work.
Authors and Affiliations
- Department of Animal Breeding and Genetics, Swedish University of Agricultural Sciences, SE-75124 Uppsala, Sweden,
Lisa S. Andersson, Doreen Schwochow, Lisbeth Wellbring, Sofia Mikko, Gabriella Lindgren & Leif Andersson - Department of Neuroscience, Uppsala University, SE-75124 Uppsala, Sweden,
Martin Larhammar, Fatima Memic, Hanna Wootz, Kalicharan Patra, Anna Vallstedt & Klas Kullander - Department of Medical Biochemistry and Microbiology, Uppsala University, SE-75123 Uppsala, Sweden,
Carl-Johan Rubin, Göran Hjälm, Freyja Imsland & Leif Andersson - Faculty of Land and Animal Resources, The Agricultural University of Iceland, IS-311 Borgarnes, Iceland,
Thorvaldur Arnason - College of Veterinary Medicine, University of Minnesota, St Paul, 55108, Minnesota, USA
Jessica L. Petersen, Molly E. McCue & James R. Mickelson - Department of Veterinary Integrative Biosciences, College of Veterinary Medicine and Biomedical Sciences, Texas A&M University, College Station, 77483, Texas, USA
Gus Cothran - Department of Bioengineering and Therapeutic Sciences, University of California San Francisco, San Francisco, 94143, California, USA
Nadav Ahituv - Institute for Human Genetics, University of California San Francisco, San Francisco, 94143, California, USA
Nadav Ahituv - Unit of Equine Studies, Swedish University of Agricultural Sciences, Uppsala, SE-75007 Sweden,
Lars Roepstorff
Authors
- Lisa S. Andersson
You can also search for this author inPubMed Google Scholar - Martin Larhammar
You can also search for this author inPubMed Google Scholar - Fatima Memic
You can also search for this author inPubMed Google Scholar - Hanna Wootz
You can also search for this author inPubMed Google Scholar - Doreen Schwochow
You can also search for this author inPubMed Google Scholar - Carl-Johan Rubin
You can also search for this author inPubMed Google Scholar - Kalicharan Patra
You can also search for this author inPubMed Google Scholar - Thorvaldur Arnason
You can also search for this author inPubMed Google Scholar - Lisbeth Wellbring
You can also search for this author inPubMed Google Scholar - Göran Hjälm
You can also search for this author inPubMed Google Scholar - Freyja Imsland
You can also search for this author inPubMed Google Scholar - Jessica L. Petersen
You can also search for this author inPubMed Google Scholar - Molly E. McCue
You can also search for this author inPubMed Google Scholar - James R. Mickelson
You can also search for this author inPubMed Google Scholar - Gus Cothran
You can also search for this author inPubMed Google Scholar - Nadav Ahituv
You can also search for this author inPubMed Google Scholar - Lars Roepstorff
You can also search for this author inPubMed Google Scholar - Sofia Mikko
You can also search for this author inPubMed Google Scholar - Anna Vallstedt
You can also search for this author inPubMed Google Scholar - Gabriella Lindgren
You can also search for this author inPubMed Google Scholar - Leif Andersson
You can also search for this author inPubMed Google Scholar - Klas Kullander
You can also search for this author inPubMed Google Scholar
Contributions
L.S.A., S.M., G.L. and L.W. collected the horse material and/or performed the genome-wide association analysis. L.S.A., D.S., M.L., G.H. and L.A. planned, designed, performed and/or analysed horse experiments. M.L., F.M., H.W., K.P., A.V. and K.K. planned, designed performed and/or analysed mouse experiments. C.-J.R. performed bioinformatic analysis. T.A. analysed horse performance data. N.A., F.I., J.L.P., M.E.M., J.R.M. and G.C. contributed with materials. L.R. recorded horse gaits. L.A. led positional cloning and characterisation of horse DMRT3. K.K. led the mouse studies. K.K. and L.A. wrote the paper with contributions from all authors.
Corresponding author
Correspondence toLeif Andersson.
Ethics declarations
Competing interests
A patent application has been filed concerning the commercial use of some of the results presented in this paper.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-7, Supplementary Methods, Supplementary Notes, Supplementary References and legends for Supplementary Movies 1-4. (PDF 14215 kb)
Supplementary Tables
This file contains Supplementary Tables 1-6. (PDF 693 kb)
Supplementary Movie 1
This movie shows a side view of a heterozygous (CA) Swedish Standardbred trotter that performs a fairly symmetric and normal trot stride as well as a very asymmetric stride (see Supplementary Information file for full legend). (MOV 10353 kb)
Supplementary Movie 2
This movie shows a side-by-side view of wild-type and _Dmrt3_-/- mice (KO) mice during swimming - see Supplementary Information file for full legend. (MOV 11867 kb)
Supplementary Movie 3
This movie shows wild-type and _Dmrt3_-/- mice during treadmill locomotion - see Supplementary Information file for full legend. (MOV 36178 kb)
Supplementary Movie 4
This movie shows wild-type and _Dmrt3_-/- mice during airstepping - see Supplementary Information file for full legend. (MOV 24035 kb)
PowerPoint slides
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/).
About this article
Cite this article
Andersson, L., Larhammar, M., Memic, F. et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice.Nature 488, 642–646 (2012). https://doi.org/10.1038/nature11399
- Received: 18 April 2012
- Accepted: 05 July 2012
- Published: 29 August 2012
- Issue Date: 30 August 2012
- DOI: https://doi.org/10.1038/nature11399