Structure of the thermally stable Zika virus (original) (raw)
Main
Zika virus (ZIKV), a flavivirus, is thought to be principally transmitted to humans by the mosquito (Aedes aegypti) vector. Other flaviviriuses include West Nile virus (WNV), Japanese encephalitis virus (JEV), dengue virus (DENV) and yellow fever virus (YFV). ZIKV generally causes a mild disease. However, when pregnant women are infected with ZIKV, there is an increased risk of developing microcephaly in the fetus1.
Retrospective analysis of data collected from a ZIKV outbreak in French Polynesia in 2013–2014 showed association of the virus with microcephaly[7](/articles/nature17994#ref-CR7 "Cauchemez, S. et al. Association between Zika virus and microcephaly in French Polynesia, 2013–15: a retrospective study. Lancet http://dx.doi.org/10.1016/S0140-6736(16)00651-6
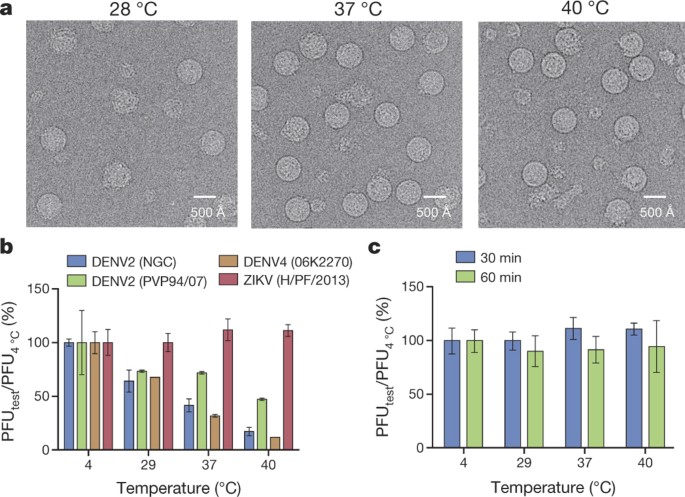
(2016)"). Here we present the 3.7 Å resolution structure of ZIKV strain H/PF/2013 isolated during that outbreak[8](/articles/nature17994#ref-CR8 "Baronti, C. et al. Complete coding sequence of Zika virus from a French polynesia outbreak in 2013. Genome Announc. 2, e00500-14 (2014)").For cryo-electron microscopy (cryoEM) studies, ZIKV was grown in the mosquito cell line at 28 °C and purified at 4 °C by polyethylene glycol precipitation, a sucrose cushion, followed by a potassium tartrate gradient. The gel analysis of the purified sample suggested it contained mostly mature virus (Extended Data Fig. 1). The ZIKV samples were incubated at 28 °C, 37 °C and 40 °C (mimicking high fever) for 30 min, before imaging by cryoEM (Fig. 1a). At 28 °C, there were broken and shrivelled particles together with some smooth surfaced particles (about 500 Å in diameter), similar to the compact DENV mature particles. Conversely, samples incubated at 37 °C and 40 °C showed many more smooth surfaced particles. The presence of a larger fraction of shrivelled particles at 28 °C could be due to the exposure of particles to high osmolality during purification. We speculate that ZIKV particles may expand into smooth surfaced particles when incubated at higher temperatures, making the lipid envelope more fluid, and allowing the structure to revert to its normal state. Some strains of DENV2 (New Guinea C, NGC) undergo notable structural changes from smooth to bumpy surfaced particles when incubated at 37 °C (refs 9, 10), whereas others (PVP94/07)11 do not. Structurally, DENV4 is considered more stable than DENV2 (NGC strain), as it remains as a smooth surfaced particle even at 40 °C, although considerable aggregation of particles occurs3. ZIKV seemed to be even more stable than DENV4, as the smooth surfaced particles did not aggregate at 40 °C (Fig. 1a). This is consistent with the plaque assay results with ZIKV, DENV2 strains (NGC and PVP94/07) and DENV4 that were pre-incubated at 28 °C, 37 °C and 40 °C for 30 min. ZIKV retained its infectivity, whereas the infectivity of DENV2 strains and DENV4 is significantly reduced at higher temperatures (Fig. 1b). The infectivity of ZIKV was only slightly reduced when the pre-incubation time was increased to 60 min (Fig. 1c).
Figure 1: ZIKV particles are more stable than DENV.
a, CryoEM micrographs of ZIKV particles incubated at different temperatures. b, ZIKV infection in BHK-21 cells is not affected when the virus is pre-incubated for 30 min at different temperatures. By contrast, all DENV strains have reduced infectivity with increasing temperatures. The y axis shows the percentage of plaque-forming units (PFU) at different temperatures divided by that of the same virus at 4 oC. c, Increased incubation time slightly reduced ZIKV infectivity. Blue and green bars indicate 30- and 60-min incubation times, respectively. Two experiments were done, and each condition was tested in triplicate. Data are mean and s.d.; means were calculated by averaging the triplicates within an experiment, and the s.d. shows the variability between the means from two separate experiments.
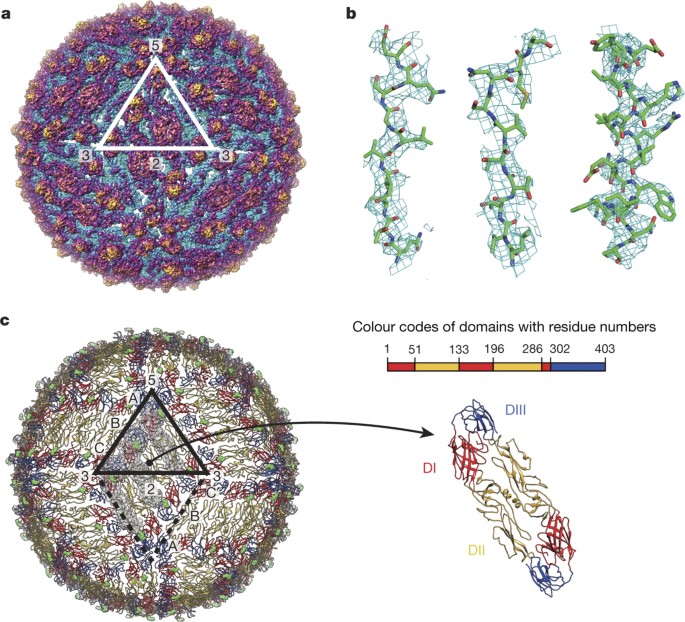
The cryoEM structure was reconstructed from the virus sample incubated at 40 °C for 30 min. The 3.7 Å resolution cryoEM map (Fig. 2a and Extended Data Fig. 2) showed well-resolved densities corresponding to the α-helices and β-strands. In addition, many side-chain densities were visible (Fig. 2b). The overall structural architecture of ZIKV (Fig. 2c) is similar to that of DENV (refs 3, 12, 13, 14) and WNV (ref. 15). While our paper was under review, a similar ZIKV structure was published[16](/articles/nature17994#ref-CR16 "Sirohi, D. et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Science http://dx.doi.org/10.1126/science.aaf5316
(2016)") showing similar features. There are 180 copies of each of the E and associated M (‘membrane’) proteins on the virus surface; they are anchored in the bilayer lipid membrane via their transmembrane regions. The E protein is the major protein involved in receptor binding and fusion. The M protein is a small protein that is hidden under the E protein layer. The E and M proteins are organized in icosahedral symmetry consisting of 60 repeating units, and each asymmetric unit contains three individual E proteins (molecules A, B and C) ([Fig. 2c](/articles/nature17994#Fig2)). The E proteins are arranged as dimers, with three dimers lying parallel to each other forming a raft; there are 30 such rafts covering the viral surface. The E ectodomain contains three domains: DI, DII and DIII ([Fig. 2c](/articles/nature17994#Fig2), right). DI acts as a bridge between DII and DIII. The tip of DII contains the fusion loop, which interacts with the endosomal membrane during fusion. The DI–DII hinge is thought to be important for flipping out DII to expose the fusion loop during the fusion event. The DI–DIII hinge is thought to be rigid at neutral pH. In most other flaviviruses, DIII contains the receptor-binding site[17](/articles/nature17994#ref-CR17 "Rey, F. A., Heinz, F. X., Mandl, C., Kunz, C. & Harrison, S. C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375, 291–298 (1995)") and has an important role in fusion[18](/articles/nature17994#ref-CR18 "Bressanelli, S. et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23, 728–738 (2004)"). After the carboxy-terminal end of the E ectodomain is the α-helical stem region that lies on the viral membrane, and the α-helical transmembrane region.Figure 2: The 3.7 Å resolution cryoEM structure of ZIKV.
a, The ZIKV cryoEM map. The map is coloured radially: 0–219 Å (cyan), 220–240 Å (purple) and >240 Å (gold). b, Zoomed-in view of the density map (cyan mesh) fitted with the E protein structure. c, The E protein arrangement on the ZIKV surface. The Asn154 glycosylation sites are shown as green spheres. The three individual E proteins (A, B and C) in one asymmetric unit (black triangle) are indicated by transparent surfaces.
The superposition of the three individual E protein molecules (A, B and C) in the asymmetric unit of ZIKV showed high similarity (Extended Data Fig. 3a), typical for flaviviruses (Extended Data Fig. 3b). Superposition of DI of molecule A of the ZIKV E protein with that of DENV2 (ref. 14) showed a slight difference in the angle at the DI–DIII and DI–DII hinges (Extended Data Fig. 3c).
Sequence comparison shows that the ZIKV E protein has similar sequence homology levels (ranging from 40 to 58%) to other flaviviruses (Extended Data Fig. 4a). There are some insertions, deletions and mutations on the E proteins that are distinct among the neurovirulent viruses (WNV, JEV and tick-borne encephalitis virus (TBEV)) and the febrile-illness-causing viruses (YFV and DENV) (Extended Data Fig. 4b). However, their contributions to the resultant pathology are not clear. A superposition of our ZIKV cryoEM structure onto the DENV2 cryoEM14 structure, and the crystal structures of WNV19 and JEV20 E proteins show structural similarities (Extended Data Fig. 5), which are consistent with the sequence comparisons (Extended Data Fig. 4b). For example, ZIKV is similar to DENV at the hi-loop of DII (Extended Data Figs 4b and 5a), where both have a three-residue long insertion compared to WNV, TBEV and JEV. However, at other sites, ZIKV is more similar to the neurovirulent JEV and WNV (Extended Data Figs 4b and 5b). For example, sequence comparison shows an Asn67Asp mutation in JEV, WNV and ZIKV compared to DENV, indicating that the glycosylation site is absent (Fig. 2c). Therefore, there is only one Asn154 glycosylation site in an E protein in ZIKV, whereas DENV has two (Asn67 and Asn153)14. The Asn154 glycosylation site is suggested to be a neurovirulence determinant in WNV21; however, DENV also has this glycosylation site but infection with DENV rarely results in neurovirulence. The Asn67 (but not Asn153) glycosylation site in DENV has been shown to be crucial for binding to DC-SIGN (also known as CD209)22 on dendritic cells. ZIKV, which also interacts with DC-SIGN23, does not have this glycosylation site, and therefore could have a different binding mechanism.
A longer glycan loop at DI is observed in ZIKV, WNV and JEV compared with TBEV, YFV and DENV1–4 (Extended Data Figs 4b and 5b). An extra residue is present in the DII kl-loop of WNV, TBEV, YFV and ZIKV compared to DENV1–4. A residue insertion at the DIII C strand in WNV, JEV, TBEV and ZIKV was observed as opposed to DENV1–4. Finally, there is a two-residue insertion at the DIII DE-loop in WNV, JEV and ZIKV compared to TBEV, YFV and DENV1–4 (Extended Data Figs 4b and 5b).
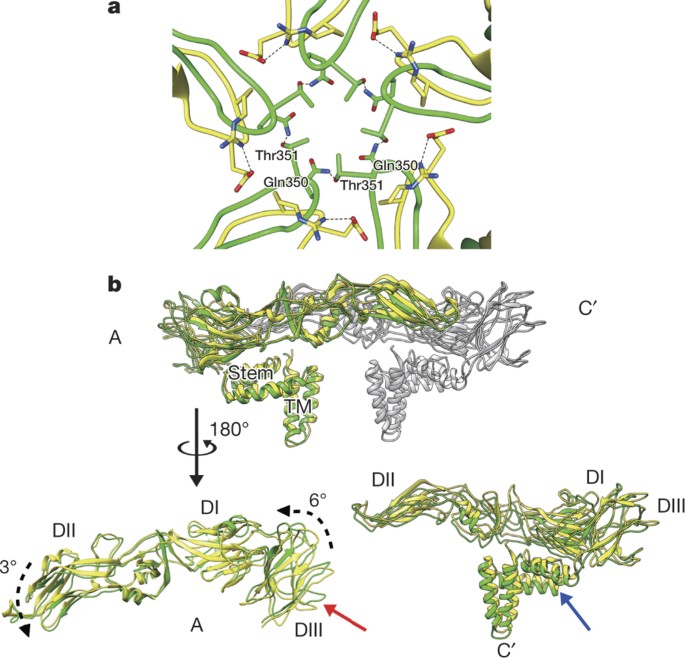
Notably, our ZIKV structure shows that the one-residue insertion at the end of the C strand of DIII (Extended Data Fig. 4b) compared to DENV may increase the overall stability of the virus. For comparison, a similar resolution DENV2 cryoEM structure14, which has been shown to be less stable at increased temperatures9,10, was used. The Ala340 insertion in the C strand of DIII allows the CD-loop to stretch further towards the five-fold vertex compared to DENV2 (Fig. 3a). Gln350 of the CD-loop from one DIII could potentially form a hydrogen bond with Thr351 from another CD-loop in the neighbouring DIII, and thus may create a hydrogen bond network between the five DIIIs (Fig. 3a and Extended Data Fig. 6). In comparison, in DENV2, there is a lack of interaction between the DIIIs in this region (Fig. 3a). The possible hydrogen bond network around the five-fold vertex of ZIKV could be responsible for the rotation of the E protein molecule A slightly (6°) anticlockwise around the axis parallel to the viral membrane (Fig. 3b, bottom left). When compared to DENV2, the position of ZIKV molecule A was at a slightly higher radius, whereas DII dips slightly closer to the viral membrane. The stem and transmembrane regions showed no notable positional differences in molecule A between DENV2 and ZIKV (Fig. 3b, top). The ectodomain of molecule C′, which forms a dimer with molecule A, is positioned at a lower radius than DENV2. The stem and transmembrane regions (Fig. 3b, bottom right), along with the membrane, are also pressed downward. The ectodomains of the B–B′ dimer also lie at a slightly lower radius (Extended Data Fig. 7b), although no positional difference in the stem and transmembrane regions was observed when compared to DENV2 (Extended Data Fig. 7a, bottom). These findings suggest that the ZIKV has a more compact surface than DENV2.
Figure 3: Possible interactions between the ZIKV DIII CD-loops around the five-fold vertex may cause differences in the position of A–C′ dimer compared to DENV2.
a, The CD-loops of ZIKV (green) are stretched closer towards the five-fold vertex than those in DENV2 (yellow). In ZIKV, the Gln350 residue may form a hydrogen bond (dashed line) to the Thr351 from the CD-loop of a neighbouring DIII, creating a hydrogen bond network around the five-fold vertex. The DENV2 residues in the CD-loop (yellow), in comparison, do not interact with other neighbouring CD-loops. b, Positional differences of ZIKV A–C′ dimer compared to DENV2. Top panel shows the side view. ZIKV and DENV molecule A are coloured green and yellow, respectively. Molecule C′ of both ZIKV and DENV2 is coloured grey. Bottom left panel show molecule A rotated by 180o to an axis perpendicular to the viral membrane. The ZIKV CD-loop (red arrow) is lifted upwards probably owing to the five-fold interaction as demonstrated in a; this molecule rotates slightly. Bottom right panel shows molecule C′ in the same orientation as in the top panel. The whole ZIKV E protein (green) molecule C′ is positioned at a lower radius than in DENV2 (yellow). The positions of the ZIKV stem (blue arrow) and transmembrane (TM) regions suggest that the membrane is pushed downwards compared to DENV2.
The E proteins on the surface of some DENV2 strains are shown to undergo structural rearrangements at 37 °C (ref. 9). In this structure, the A–C′ dimer moves outwards to a higher radius and also rotates slightly. The B–B′ dimer moves to a higher radius and the E proteins dissociate from each other. A study of an antibody, which traps a transitional stage of this motion11, suggests that the A–C′ dimer probably moves to a higher radius before the B–B′ dimer11. In the ZIKV structure, the possible network of interactions around the five-fold vertex between the five A–C′ dimers may result in tighter packing that prevents this structural transition.
Both the TIM (T cell, immunoglobulin and mucin) and TAM (Tyro3, Axl and Mer) phosphatidylserine transmembrane receptors have been shown to be used by DENV for cell entry24. Conversely, ZIKV was shown to bind to TAM but not TIM receptors23. It is possible that the looser surface of DENV2 compared to ZIKV could allow more interactions with a wider range of phosphatidylserine-binding receptors.
Unlike DENV, ZIKV is detected in the urine6, saliva5 and semen4 (which allows potential transmission by sexual contact25), suggesting that the virus is stable enough to withstand harsh conditions. This is consistent with our infection assay showing that ZIKV is more thermally stable than DENV. Furthermore, the potential interactions identified in our structure may explain the stability of ZIKV and may serve as potential sites to exploit for future therapeutics.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Virus plaque assay to determine the stability of ZIKV and DENV serotypes at different temperatures
The virus strains used for the plaque assay experiments were: DENV2 (NGC), DENV2 (PVP94/07), DENV4 (06K2270) and ZIKV (H/PF/2013). Virus samples were pre-incubated for 30 min at 4 °C, 29 °C, 37 °C and 40 °C and chilled at 4 °C for at least 30 min. Samples were serially diluted and added to BHK-21 (ATCC) cells grown in 24-well plates. For plaque assays, the cells were not specially authenticated, and not tested specifically for mycoplasma contamination. Virus was incubated with the cells for 1 h at 37 °C, then layered with methyl cellulose. After incubation at 37 °C for 4 days, the cells were fixed and stained to determine the plaque-forming units.
A similar experiment was done by pre-incubating ZIKV H/PF/2013 for 60 min at the same temperatures, before adding to BHK-21 cells.
Virus sample preparation
ZIKV strain H/PF/2013 was grown in Aedes albopictus C6/36 cells (ATCC) supplemented with RPMI1640 medium and 2% fetal bovine serum. The cells were inoculated with ZIKV at a multiplicity of infection of 2, and incubated at 29 °C for 3 days. Virus-containing medium was subjected to centrifugation at 12,227_g_ for 45 min to remove cells and other debris, and the virus was precipitated with 8% (w/v) polyethylene glycol 8000 in NTE buffer (10 mM Tris-HCl, pH 8.0, 120 mM NaCl and 1 mM EDTA) overnight. Virus was pelleted by centrifugation at 14,334_g_, resuspended in NTE buffer and purified through a 24% sucrose cushion. Virus was further purified by ultracentrifugation using a 10–30% (w/v) potassium tartrate gradient. Virus band was extracted from the gradient with a syringe, and then the sample was buffer-exchanged into NTE buffer and concentrated using Amicon Ultra-4 100-kDa centrifugal concentrator (Millipore). Virus concentration and purity was estimated by Coomassie-blue-stained SDS–PAGE gel, where different concentrations of bovine serum albumin (BSA) solution were used as standards. All steps of the purification procedure were done at 4 °C.
CryoEM sample preparation
Virus samples were incubated at 4 °C, 29 °C, 37 °C or 40 °C, for 30 min, then kept at 4 °C before freezing on cryoEM grids. About 2 μl of sample was put on a copper cryo-EM grid (with lacey carbon covered with thin carbon film), blotted for 1 s and plunged in liquid ethane bath using the Vitrobot plunger (FEI). The grids were stored in liquid nitrogen.
CryoEM data collection, image processing and 3D reconstruction
Images of vitrified virus were collected on a Titan Krios (FEI) microscope at 300 kV in nanoprobe mode using Leginon software for automated data collection. The magnification was 104,477, giving a pixel size of 1.34 Å. The images were recorded on Falcon II direct electron detector (FEI) in movie mode, with 30 frames per 1.8 s exposure time, giving a total dose of 50 e− Å−2. The individual frames were mutually aligned to correct for beam-induced sample drift with the MotionCorr program26. The first 10 frames were combined to produce the average image for further processing. The higher contrast full dose average was also calculated and used for defocus determination and for manual particle picking. In total, 11,600 individual virus particles from 3,083 micrographs were selected. The 3D reconstruction was carried out with MPSA27, by using a 3D map of DENV4 (EMD-2485) as the starting model, with icosahedral symmetry averaging. The image data was split into two halves and processed with MPSA independently, to follow the ‘gold standard’ procedure for resolution determination. After 20 cycles of orientation search and 3D reconstruction, the final map was produced by using 7,180 particle images. The resolution of the map is 3.7 Å, at Fourier shell correlation cut off of 0.143 (Extended Data Fig. 2).
Model building and refinement
The resolution of the map allowed us to trace all of the polypeptide chains: three E and three M proteins. After manual structure building in Coot28, using DENV2 coordinates (PDB 3J27) as a starting template, modified to include insertions, the protein structure had been refined with CNS29 with icosahedral symmetry constraints applied, and showed _R_work and _R_free values of 37% and 39%, respectively. For refinement, the map has been ‘sharpened’ with application of a B-factor of −55 and low-pass filtered to 3.7 Å. Additional secondary structure restrains were applied to avoid overfitting. After refinement, the fit and the geometry of the protein structure was checked using Coot tools.
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
The cryoEM 3D reconstruction map of ZIKV has been deposited with the Electron Microscopy Data Bank (EMDB) under accession code EMD-8139. The modelled structure of the virus glycoprotein shell has been deposited with the Protein Data Bank (PDB) under the accession code 5IZ7.
References
- Schuler-Faccini, L. et al. Possible association between Zika virus infection and microcephaly – Brazil, 2015. MMWR Morb. Mortal. Wkly. Rep. 65, 59–62 (2016)
Article Google Scholar - Cao-Lormeau, V. M. et al. Guillain–Barre syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387, 1531–1539 (2016)
Article Google Scholar - Kostyuchenko, V. A., Chew, P. L., Ng, T. S. & Lok, S. M. Near-atomic resolution cryo-electron microscopic structure of dengue serotype 4 virus. J. Virol. 88, 477–482 (2014)
Article Google Scholar - Mansuy, J. M. et al. Zika virus: high infectious viral load in semen, a new sexually transmitted pathogen? Lancet Infect. Dis. 16, 405 (2016)
Article Google Scholar - Barzon, L. et al. Isolation of infectious Zika virus from saliva and prolonged viral RNA shedding in a traveller returning from the Dominican Republic to Italy, January 2016. Euro Surveill. 21, 1 (2016)
Google Scholar - Gourinat, A. C., O’Connor, O., Calvez, E., Goarant, C. & Dupont-Rouzeyrol, M. Detection of Zika virus in urine. Emerg. Infect. Dis. 21, 84–86 (2015)
Article CAS Google Scholar - Cauchemez, S. et al. Association between Zika virus and microcephaly in French Polynesia, 2013–15: a retrospective study. Lancethttp://dx.doi.org/10.1016/S0140-6736(16)00651-6 (2016)
- Baronti, C. et al. Complete coding sequence of Zika virus from a French polynesia outbreak in 2013. Genome Announc. 2, e00500-14 (2014)
Article Google Scholar - Fibriansah, G. et al. Structural changes in dengue virus when exposed to a temperature of 37 degrees C. J. Virol. 87, 7585–7592 (2013)
Article CAS Google Scholar - Zhang, X. et al. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl Acad. Sci. USA 110, 6795–6799 (2013)
Article CAS ADS Google Scholar - Fibriansah, G. et al. DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 349, 88–91 (2015)
Article CAS ADS Google Scholar - Kostyuchenko, V. A., Zhang, Q., Tan, J. L., Ng, T. S. & Lok, S. M. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J. Virol. 87, 7700–7707 (2013)
Article CAS Google Scholar - Kuhn, R. J. et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108, 717–725 (2002)
Article CAS Google Scholar - Zhang, X. et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nature Struct. Mol. Biol. 20, 105–110 (2013)
Article Google Scholar - Mukhopadhyay, S., Kim, B. S., Chipman, P. R., Rossmann, M. G. & Kuhn, R. J. Structure of West Nile virus. Science 302, 248 (2003)
Article CAS Google Scholar - Sirohi, D. et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Sciencehttp://dx.doi.org/10.1126/science.aaf5316 (2016)
- Rey, F. A., Heinz, F. X., Mandl, C., Kunz, C. & Harrison, S. C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375, 291–298 (1995)
Article CAS ADS Google Scholar - Bressanelli, S. et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23, 728–738 (2004)
Article CAS Google Scholar - Nybakken, G. E., Nelson, C. A., Chen, B. R., Diamond, M. S. & Fremont, D. H. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 80, 11467–11474 (2006)
Article CAS Google Scholar - Luca, V. C., AbiMansour, J., Nelson, C. A. & Fremont, D. H. Crystal structure of the Japanese encephalitis virus envelope protein. J. Virol. 86, 2337–2346 (2012)
Article CAS Google Scholar - Beasley, D. W. et al. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J. Virol. 79, 8339–8347 (2005)
Article CAS Google Scholar - Pokidysheva, E. et al. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell 124, 485–493 (2006)
Article CAS Google Scholar - Hamel, R. et al. Biology of Zika virus infection in human skin cells. J. Virol. 89, 8880–8896 (2015)
Article CAS Google Scholar - Meertens, L. et al. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12, 544–557 (2012)
Article CAS Google Scholar - McCarthy, M. US health officials investigate sexually transmitted Zika virus infections. Br. Med. J. 352, i1180 (2016)
Article Google Scholar - Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584–590 (2013)
Article CAS Google Scholar - Liu, X., Jiang, W., Jakana, J. & Chiu, W. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J. Struct. Biol. 160, 11–27 (2007)
Article CAS Google Scholar - Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Article CAS Google Scholar - Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Article Google Scholar
Acknowledgements
We thank the European Virus Archive for providing us with the ZIKV H/PF/2013 strain. We thank S. Lambert for helping to edit the paper. The work is supported by Singapore Ministry of Education Tier 3 grant (MOE2012-T3-1-008), National Research Foundation Investigatorship award (NRF-NRFI2016-01) awarded to S.-M.L. and the Duke-NUS Signature Research Programme funded by the Ministry of Health, Singapore.
Author information
Author notes
- Victor A. Kostyuchenko, Elisa X. Y. Lim and Shuijun Zhang: These authors contribute equally to this work.
Authors and Affiliations
- Program in Emerging Infectious Diseases, Duke–National University of Singapore Medical School, Singapore, 169857, Singapore
Victor A. Kostyuchenko, Elisa X. Y. Lim, Shuijun Zhang, Guntur Fibriansah, Thiam-Seng Ng, Justin S. G. Ooi & Shee-Mei Lok - Department of Biological Sciences, Centre for BioImaging Sciences, National University of Singapore, Singapore, 117557, Singapore
Victor A. Kostyuchenko, Elisa X. Y. Lim, Shuijun Zhang, Guntur Fibriansah, Thiam-Seng Ng, Justin S. G. Ooi, Jian Shi & Shee-Mei Lok - Department of Biological Sciences, CryoEM unit, National University of Singapore, Singapore, 117557, Singapore
Jian Shi
Authors
- Victor A. Kostyuchenko
- Elisa X. Y. Lim
- Shuijun Zhang
- Guntur Fibriansah
- Thiam-Seng Ng
- Justin S. G. Ooi
- Jian Shi
- Shee-Mei Lok
Contributions
E.X.Y.L. prepared the ZIKV sample for cryoEM work and carried out the plaque assays for testing the thermal stability of ZIKV and DENV. E.X.Y.L. and T.-S.N. optimized conditions for freezing samples on cryoEM grids. J.S. and T.-S.N. collected cryoEM images. G.F., S.Z. and J.S.G.O. boxed particles for cryoEM image reconstruction. T.-S.N. checked CTF curves of the micrographs. V.A.K. optimized and carried out the cryoEM image reconstruction procedure. G.F., S.Z. and V.A.K. fitted the structure into the cryoEM map. G.F., S.Z., V.A.K. and S.-M.L. analysed the structure. G.F., S.Z. and S.-M.L. prepared the figures. S.-M.L. and V.A.K. designed experiments. G.F., V.A.K., J.S., E.X.Y.L. and S.-M.L. wrote the manuscript. All authors discussed results and commented on the manuscript.
Corresponding author
Correspondence toShee-Mei Lok.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks J. Huiskonen and Y. Modis for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 The Coomassie-blue-stained SDS–PAGE gel profile of ZIKV.
The virus preparation showed clear protein bands corresponding to the E protein (50 kDa), capsid (C) protein (15 kDa), and M protein (10 kDa). The premembrane (prM) band (25 kDa) is not detected, indicating that there is little contamination by immature virus particles.
Extended Data Figure 3 Comparison of ZIKV with DENV2 E proteins.
a, Superposition of ZIKV E protein molecules A, B and C using DI as a reference point. The molecules are similar to each other. b, Superposition of the three individual DENV2 E proteins in an asymmetric unit by their DIs show high similarities between them. c, Superposition of molecule A of ZIKV with that of DENV2 by their DIs shows that they have slightly different DI–DIII and DI–DII hinge angles.
Extended Data Figure 4 Sequence comparison of ZIKV to neurovirulent viruses (WNV, JEV and TBEV) and to febrile-illness-causing viruses (YFV and DENV1–4).
a, b, Percentage sequence similarity (a) and sequence alignment (b) of flaviviruses. Green boxes indicate the most different regions between the neurovirulent viruses and the febrile-illness-causing viruses. Contributions of these regions to the virus pathology are unknown. ZIKV contains regions that are distinct in both virus groups.
Extended Data Figure 5 Structural comparison of different regions on the E protein sites unique to febrile-illness-causing virus and neurovirulant virus to ZIKV.
a, Comparison of the hi-loop between ZIKV and DENV by superimposing their DIIs. b, Comparison of the glycan loop (DI), CD-loop (DIII) and DE-loop (DIII), between ZIKV, WNV and JEV by superimposing their DIIIs. Comparison of the kl-loop (DI–DII hinge) by superimposing these viruses by DI.
Extended Data Figure 6 The fitted CD-loop in the cryoEM density around the five-fold vertex.
Extended Data Figure 7 The ectodomain of the ZIKV B–B′ dimer is positioned lower in the radius compared to DENV2.
a, Top panel shows the top view of the B–B′ dimer of ZIKV (green) and DENV2 (yellow), while the bottom panel shows the side view. The position of stem and transmembrane regions in both ZIKV and DENV are similar. b, The ZIKV E ectodomain is positioned at a slightly lower radius than DENV2. Stick connecting similar atoms in ZIKV and DENV is shown to help with the comparison of the positions of the E ectodomains.
PowerPoint slides
Rights and permissions
About this article
Cite this article
Kostyuchenko, V., Lim, E., Zhang, S. et al. Structure of the thermally stable Zika virus.Nature 533, 425–428 (2016). https://doi.org/10.1038/nature17994
- Received: 24 March 2016
- Accepted: 11 April 2016
- Published: 19 April 2016
- Issue Date: 19 May 2016
- DOI: https://doi.org/10.1038/nature17994