Deep resequencing reveals excess rare recent variants consistent with explosive population growth (original) (raw)
Introduction
Models of human genetic diversity have an important role in both population genetics and genetic epidemiology, and considerable effort has been expended to characterize human genetic diversity by directly sequencing DNA from large population samples (for example, refs 1, 2, 3, 4) and from the ongoing 1000 Genomes and Personal Genomes projects[5](/articles/ncomms1130#ref-CR5 "1000 Genomes. A deep catalog of human genetic variation, http://www.1000genomes.org
(2010)."),[6](/articles/ncomms1130#ref-CR6 "Personal Genome Project.
http://personalgenomes.org
(2010)."). To study the distribution and role of truly rare variants, it is necessary to sequence extremely large numbers of individuals, a project that has only recently become feasible.An inherent problem with such a deep resequencing effort is distinguishing actual rare genetic variants from stochastic sequencing errors, which will occur at almost every site if enough individuals are resequenced. Regenotyping the sites of potential rare variants can mitigate this uncertainty; however, for large-scale studies, there are so many rare single-nucleotide polymorphism (SNP) calls that regenotyping all of them becomes cost prohibitive. Another approach is to focus on variants for which the sequence evidence is very strong. This worked well for Sanger sequencing in the HapMap project7; however, crucially, the HapMap project had a much smaller sample size and targeted high-confidence calls of common SNPs for inclusion on a SNP chip. False-negative SNP calls were a minor concern for HapMap, whereas for a comprehensive catalogue of rare variation, it is important to minimize them. Therefore, to catalogue rare variants in a thorough and cost-effective manner, in this study we assign probabilities to genotype calls, explicitly estimating our uncertainty for each call. This approach differs from earlier methods8,9 by using the overlapping genotypes of the (Sanger) ENCODE and (SNP Chip) HapMap projects to train a Dirichlet mixture which relates genotypes to the distribution of phred probability scores. As we assign probabilities to all genotype calls, all analyses of the genotypes are also probabilistic, carrying the genotype uncertainty forward to quantitative estimates of the resulting uncertainty in our sitefrequency spectrum (SFS) and population-genetic estimates.
In this study, we selected genes _KCNJ1_1 and HHEX for resequencing in 13,715 individuals. Such a large sample made some unique population-genetic calculations possible, such as a model of the growth rate of the European population over the last few thousand years. Earlier population-genetic models of European population growth10 based their estimates on higher-frequency variants than the bulk of variants ascertained in our study. By estimating the distribution of times at which a variant of a given contemporary frequency might have plausibly arisen in the ancestral population, we have been able to compare our growth-rate estimate with earlier estimates. We were also able to separately estimate mutation rate and demographic parameters, which are normally confounded in equilibrium population genetics11, and estimate the mutation rate specific to each gene locus, using a population-genetic model similar to those described in Wakeley and Takahashi11 and Boyko et al.12 Our SFS showed far more rare variation than would have been predicted by classical models of population genetics, and our population genetics calculations established a clear genetic signal of a recent acceleration in European population growth.
Results
Sequencing assay
We resequenced these genes in 13,715 ARIC (Atherosclerosis Risk In Communities) individuals (3,293 African Americans and 10,422 individuals of European ancestry, Supplementary Methods). We applied Sanger sequencing of PCR products from genomic DNA from 50 amplicons covering the coding regions, introns and near flanking regions (see Supplementary Methods). We achieved excellent sequence coverage of the loci, and called variant sites in a way that assigned probability scores to each (see Methods). To quantify the rate of sequencing error, we validated potential rare variants by barcoding and pooling of the relevant PCR amplicons, and then submitted them en masse to 454 Roche sequencing. We validated 216 sites, the majority of which were high-probability variant calls in our initial genotyping. We used the results of this exercise to calibrate our genotype probabilities with their accuracy in calling rare variants (Supplementary Methods).
There are many variants with predicted biological impact: combined over the two genes, there are 35 sites with a probability of at least 0.35 that the genotype varies in a way that would change the protein product; 19 of these are identified by PolyPhen13 as potentially damaging, on the basis of cross-species sequence conservation at those sites. Figure 1 is a survey of the potentially damaging variants, and gives the probabilities that the variants are present at the indicated sites. By contrast, dbSNP14 (http://www.ncbi.nlm.nih.gov/projects/SNP/) reports just two and ten missense SNPs in HHEX and KCNJ11, respectively. Although a PolyPhen call of 'damaging' is not definitive, this observation of rare variation at a large number of evolutionarily conserved sites is consistent with the expectation that rare, recent variants are more randomly distributed than common variants, because selection pressures have had less time to act on them. The KCNJ11 protein structure in Figure 1 was determined by homology modelling using SWISS-MODEL15, and covers residues 33–357 (out of a total of 390 residues). There were other protein-changing KCNJ1 variants at sites not covered by the model. Those identified by Polyphen as benign were V13M, R29H and L361F (posterior probabilities 1.00, 0.35 and 0.85, respectively). Those identified as potentially damaging were E23K, R31W, R371H, P374R and S385C (posterior probabilities 1.00, 0.36, 0.97, 0.96 and 1.00, respectively). No sufficiently homologous crystal structure for HHEX is available for homology modelling; therefore, we show the gene structure instead (Fig. 1b).
Figure 1: Physical location of selected variants.
For each variant shown, the figure shows the reference residue, the location, the variant residue and, in parentheses, the variant's posterior probability. Variants identified by Polyphen13 as potentially damaging to the protein product are shown in magenta, others are in cyan. (a) Variants that change the protein structure in KCNJ11. (b) Variants in HHEX. No sufficiently homologous crystal structure for HHEX is available for homology modelling; hence, we show the gene structure instead. Blue regions depict exons. Green regions depict neighbouring intronic/untranslated regions (30 base pairs in both directions). Black bars indicate excluded intronic sequence. Non-coding variants are shown in grey, and show the reference allele, the build 36 coordinate on chromosome 10, the variant allele and the posterior probability of the variant.
Variant counts
The data reveal a vast number of rare variants. By drawing repeated samples from the genotype probabilities, counting the number of variants in each such sample and taking the average of these counts, we compute the expected number of variant sites in this sample to be 578.6 (s.d.=7.6), with 316.9 in HHEX and 261.7 in KCNJ11 (see descriptions of loci and count sampling in Methods). In sub-samples, the number of singleton-variant sites observed increases almost linearly with sample size (Fig. 2, which shows the counts for variants found in the European-American cohort).
Figure 2: Number of variants as a function of sample size.
Counts of the number of observed segregating sites as a function of sample size for (a) HHEX and (b) KCNJ11. Solid blue line shows the total number of segregating sites. Red shows singletons, and yellow, brown and purple lines show the numbers of variants with relative minor allele frequency <0.01, 0.01–0.05 and more than 0.05, respectively. Roughness in these curves indicates stochasticity in the number of variants observed across multiple sample populations. Dashed lines show extrapolations of the expected number of segregating sites in larger samples according to Watterson's classical estimate. In all cases, we found far more segregating sites at larger sample sizes than Watterson's estimate would have predicted.
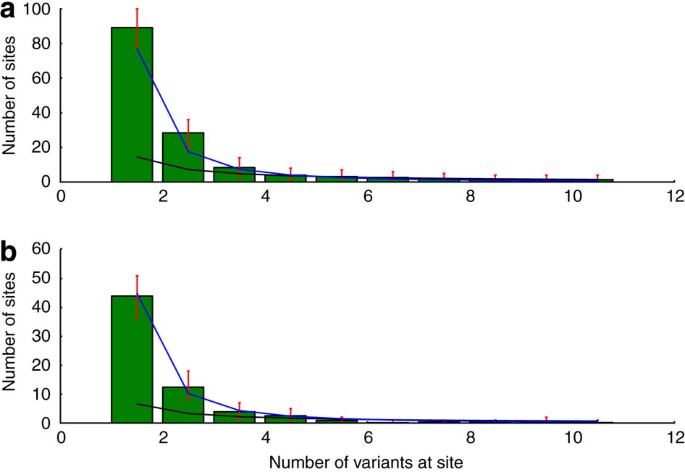
The full spectra for both population groups are shown in Supplementary Figure S1. Spectra for synonymous and non-synonymous variants are shown in Supplementary Figure S2. The spectra for non-coding regions seem to be very similar to those in Figure 3, because non-coding sites dominate the class of sites in which nucleotide variation can have no impact on the protein residue sequence.
Figure 3: Site-frequency spectra.
Site-frequency spectra in (a) HHEX and (b) KCNJ11 over 'neutral sites' (see Methods) in the two genes for the European sub-population. The x axis depicts the number of variants observed at a site; the y axis depicts the expected number of sites at which that many variants were seen. Green bars show the expected number of sites, as determined by sampling from the posterior genotypic distributions for each sampled individual, and error bars show the 99% confidence intervals from these samples. The black line shows the expected SFS spectrum, given the Wright–Fisher constant population size model and mutation rate Θ estimated by Watterson's method (Equation 4.16, Hartl & Clark (2007)) The blue line shows the mean posterior SFS given the population model used to calculate the mutation rate in Figure 4.
Population genetic calculations
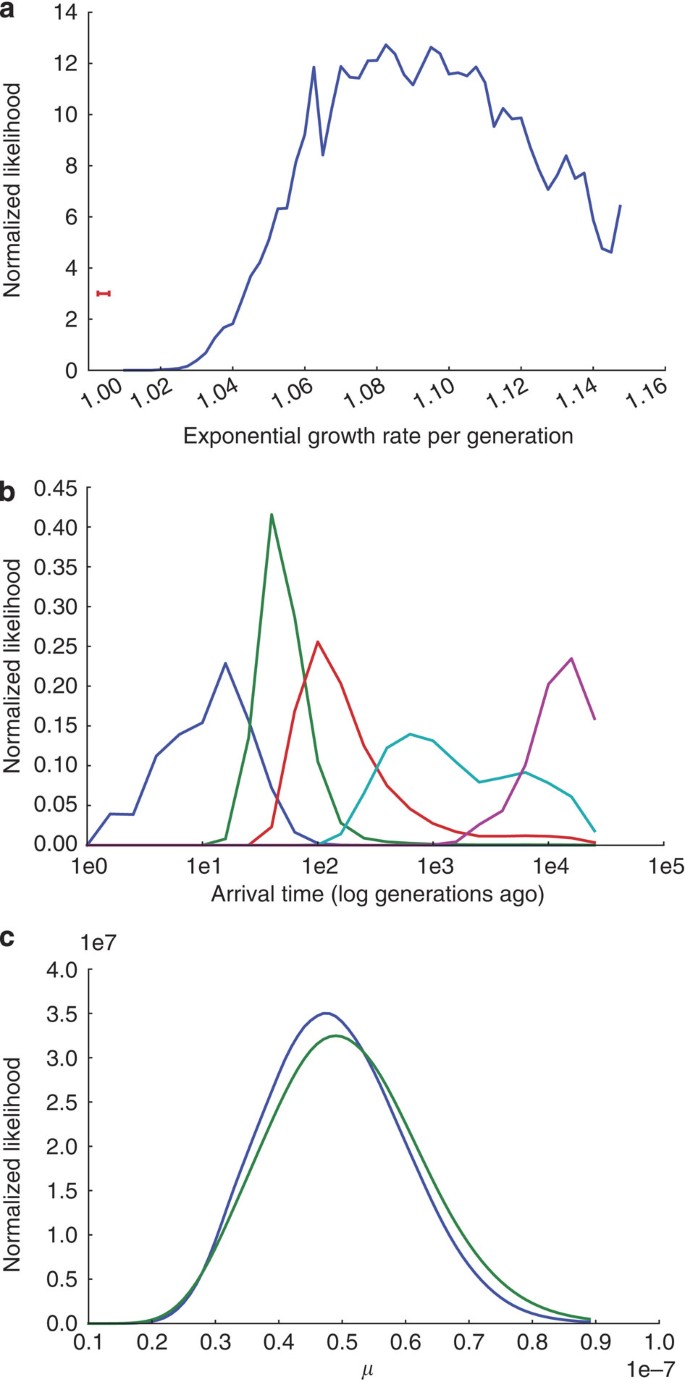
We fit a model of exponential growth to the SFS (see Fig. 3) of our European-American sample (see Methods). To avoid complications from evolutionary selection pressures, such as those described in Ohta16, we restricted this analysis to sites at which no selection pressure is to be expected (Methods). The excess of rare variants in HHEX and KCNJ11 fits well with this model (Fig. 3), giving a mean posterior growth rate of 1.094 (that is, an increase of 9.4%) per generation (Fig. 4a).
Figure 4: Mutation rate estimates.
These estimates are based on drawing an average over 100 coalescent trees per grid point. (a) Estimated marginal posterior distribution over growth rates per generation during the exponential growth phase. Red error bar in the lower left-hand corner shows the 95% confidence interval of the growth rate in the European lineage estimated in Table 1 of Gutenkunst et al.10 which is much lower, because the more common variants used in that estimate pertain to a more remote time in our history. (b) Estimated marginal posterior distributions on the time when variants of various relative minor allele frequencies arose in the population, relative to the logarithm of number of generations ago. Blue, green, red, cyan and magenta lines correspond to distributions for variants with relative minor allele frequency (RMAF) of 5×10−5, 5×10−4, 5×10−3, 5×10−2 and 5×10−1, respectively. A RMAF of 5×10−5 corresponds to singletons in our data set, which, according to our model, mostly arose in the last 2,500 years. Most previous analyses have dealt with SNPs with a RMAF on the order of 5×10−2, corresponding to much earlier mutations. (c) Estimated marginal posterior distribution over mutation rates given the SFS in the two genes. Blue and green lines are for HHEX and KCNJ11, respectively.
The variance in this estimate is high; however, combined with earlier demographic estimates, our growth-rate estimate gives a clear genetic signal that over the last few millennia, the rate of population expansion has accelerated substantially. Because previous genomic studies of human population samples have been based on either resequencing a small group of individuals or on HapMap SNPs ascertained with a bias towards common variation, these have only captured the distribution of common variants (relative minor allele frequency ∼0.05). As shown in Figure 4b, we find that most of the variants in this part of the frequency spectrum arose about 100–3,000 generations ago, or about 2,500–75,000 years ago, assuming 25 years per generation. In the exceptionally large sample resequenced here, singletons correspond to mutations that arose during the last ∼100 generations (Fig. 4b), and thus carry information about the demographics of Europe after its widespread adoption of agriculture17. Despite relying on shallower regenotyping data, earlier studies10 also found a good fit to an exponential growth model, but with the substantially lower modal growth rate of 1.004 per generation. Our posterior distribution implies that the growth rate is bound below by 1.015; hence, by comparing our results with those of Gutenkunst et al.10, we conclude that Europe's population growth rate accelerated substantially over the last 2,000 years, and our mean posterior growth-rate estimate implies an acceleration by more than an order of magnitude. Reliable census estimates of Europe's population begin around 155018, and the growth rate in Europe since 1600 has been ∼11.5% per generation (estimated from Table 1.3 of Livi-Bacci19, assuming 25 years per generation). This suggests that in future, even deeper resequencing efforts will reveal an SFS with even greater proportions of rare and missense variants with potential consequences for human health.
Mutation rate estimates
Our model yields mean mutation rate estimates of ∼4.9×10−8 and ∼5.1×10−8 mutations per site per generation for HHEX and KCNJ11, respectively (Fig. 4c). These estimates are in approximate agreement with earlier genome-wide estimates based on human–chimpanzee divergence20 or de novo mutations in monogenic disorders21; however, our extremely deep sampling has enabled estimates local to each gene. These estimates will become more precise in the future using samples across larger sets of genes (which will make the demographic parameter estimates more precise) and larger groups of people (which will make all parameter estimates more precise).
Discussion
The majority of the variants we found are extremely rare and could only have been captured by the kind of exceptionally deep resequencing described here (Figs 1 and 2). From the perspective of classical population genetics for stable populations, this abundance of rare variation is surprising: the expected number of singletons in our data is at least five times greater than the standard Wright–Fisher model22 would predict, and we find a nearly linear increase in the number of singletons as the sample size grows, whereas the classic theory for stable populations predicts that discovery of new variant sites will rapidly saturate as the sample size increases. This departure from the expected distribution suggests recent explosive human population growth, which has produced gene genealogies with a preponderance of short, recent genealogical branches. Assuming mutations are uniformly distributed over these genealogies, most mutations will have fallen on those very recent branches; thus, most variants will appear in only a few contemporary individuals. We thus predict that rare human genetic variation will tend to be more damaging than the common variants that have been the main focus of genetic studies up to this point. This is because in a sufficiently large sample, every human gene is likely to harbour many rare variants that have arisen so recently that selection can have influenced the frequencies of only the most severely deleterious alleles (Fig. 1). We also predict that all individuals carry multiple loss-of-function alleles in their genome21,23,24,25,26. Although these predictions must be tested in a future assay, a simple calculation assuming a conservative mutation rate of 1×10−9 still implies that the human genome of ∼3×109 sites is saturated with mutations arising just in the current human generation of 6.7×109 people. This supports the concerns raised by Lynch21 regarding burgeoning human “mutational load” and bears on the 'missing heritability' still unexplained by genome-wide association studies27. This myriad of rare but potentially large-effect variants are embedded in diverse gene regulatory pathways, suggesting that considerable phenotypic differences may arise from low-frequency genetic architectures28,29. Knowledge of these variants may provide good individual phenotypic predictive ability within families, but their individual rarity in a population will mean that they will have very low population-attributable risk. This suggests that the best scale for inference about the genetics of complex disorders may be individuals with genomic regions of highly shared ancestry, including the family unit itself.
Methods
DNA sequencing
Using traditional Sanger fluorescent dideoxy methods on ABI 3730 capillary sequencers, we resequenced HHEX and KCNJ11 in the ARIC cohort. Primers were designed to yield tiled amplicons across the full HHEX (7.9 kb) and KCNJ11 (5.5 kb) genes, including exons and introns and 1.2 kb upstream of the transcriptional start site, so that the coverage began for HHEX at global coordinate (build 36) 94438570 and ended at 94446433, and for KCNJ11 began at 17362320 and ended at 17367885. See Supplementary Table S1 for primer sequences. The design was successful in providing coverage for more than 90% of each gene. Low-quality sequence reads and amplicons that failed to map to the related amplicon in the human reference sequence were removed, leaving 50 amplicons in overlapping tiles across the genes. There was one region that failed to map in HHEX that spanned 442 bp, including 100 bp of the coding region. There were three such regions in KCNJ11: one spanning 43 bp, another spanning 59 bp and a third spanning 537 bp, none of which were in the coding regions. These mapping failures were likely caused by very high GC content interspersed with simple tandem repeats. Trace files have been deposited in the NCBI trace archive under center_name="bcm" and center_project="rhicf".
Genotype calling
Rare variants are a challenge to score by Sanger sequencing, because they are likely to be present in a population sample only as heterozygotes. Heterozygotes are difficult to distinguish from aberrations in Sanger sequencing data, because the PCR products in the sequencing gel come from both chromosomes; hence, heterozygous genotypes appear as a superposition of the two alleles. In particular, a heterozygous SNP resembles a 'double peak' in the Sanger trace file, but so do certain types of noise. The most popular program for analysing Sanger sequence data, phred30, was designed for haploid organisms, and explicitly treats such superposition as a sign of noisy data; hence, it cannot be used to directly identify heterozygous SNPs. Various approaches have been used to overcome this limitation by using only phred to highlight the double peaks, then analysing them separately. However, phred was carefully trained on a substantial corpus of sequencing data, although from haploid clones; therefore, we developed a way to make use of that previous training to evaluate each of the peaks by masking one allele to render our diploid sequence haploid-like (detailed below). In our approach, we effectively separated the colour channels at double peaks to obtain separate phred calls for each possible nucleotide in such a way that, as much as possible, phred would interpret them as clean haploid peaks.
The distribution of phred software comes with the source code; hence, we were able to modify the way it responds to double peaks in the trace files: we programmed it to zero out each peak channel so that the other peak could be rescored using phred's standard machinery without being confounded by the superposition. Phil Green and Brent Ewing have very generously agreed to permit the use and distribution of our patch to phred, and to continue providing the version of the source code to which it applies (version 0.020425.c.) Once modified, phred reports up to three scores for each of the peaks in a trace file: it always reports the standard phred score, and in the case of a double peak it potentially also reports the two scores obtained by zeroing out each of the peak channels. There is only one instance when these two extra scores are censored: it often happens that the dominant channel for one peak 'runs on' into an overlap with the next peak, creating a double peak that has nothing to do with genetic variation. We programmed phred to ignore a double peak if it immediately follows a peak on the same channel as its minor peak, and the amplitude of the minor peak is <20% of that of the major peak.
A number of individuals genotyped in the HapMap 3 project were also sequenced in the ENCODE project31, and we used this comparison data to train the priors in our model. We took the HapMap 3 genotypes as the gold standard, and at 830 sites with both HapMap 3 genotypes and Sanger sequence data we ran our modified phred. For each site with a given genotype g, as reported by HapMap 3, we counted how many times each type of peak was observed in the ENCODE trace files at that site. Trace files from the reverse strand were associated with the complementary genotype. This gave us a set of per-site counts for types of peaks. We used these counts to train Dirichlet mixture priors Tg (on the types of peaks observed at a site with given genotype g) and Sgp (on the phred scores observed at a site with genotype g and peak type p) in much the same way as described in Sjölander et al.32, except that we considered priors with up to six mixture components and, for each number of mixture components, we sampled mixture weights and pseudocounts using Hamiltonian Markov Chain Monte Carlo33, using a flat hyperprior on the mixture weights and exponential distributions of mean 1 on the pseudocounts for each component. From among the six priors constructed this way (one for each possible number of mixture components), we chose the one with the highest total probability (that is, the probability of the data given the constructed prior multiplied by the probability of the prior given the hyperprior) and used that as Tg. Similarly, for each genotype g and peak type p, we constructed counts for the scores associated with them as follows: for every site where this genotype and peak type was observed, we counted the number of times each phred score was observed in people with that genotype and peak type at that site. We trained a mixture prior Sgp from these counts in the same way as we trained Tg.
We used this prior in a Markov Chain Monte Carlo series at each site that also accounted for the population-level genotype frequencies at the site, so that it takes stronger evidence to call rarer genotypes. The software application, SnppnS, is available as Supplementary Software or can be downloaded from http://micortex.org/software.php.
Sampling of site-frequency spectra
Because we generate genotype probability distributions rather than categorical genotype calls, there is some uncertainty in the SFS for our genetic data. To generate Figure 3, we drew samples from the posterior genotype distributions for every position, and used these categorical genotypes to generate an SFS. We repeated this sampling 1,000 times. The error bars in Figure 3 represent the 99% confidence intervals on the site counts for each bin. To show concordance with the predicted SFS from the population genetics calculation in the next section, the spectra in Figure 3 show the frequency distribution only for variants at the same set of 'neutral' sites used in that calculation: wobble sites or those at least 30 base pairs from a coding region. So that each site would come from a population of the same size, in the European (African)-American sample, we ignored all sites called in fewer than 10,000 (3,000) people. For sites called in more than 10,000 (3,000) people, we randomly sampled a group of 10,000 (3,000) and drew our counts from that.
Population genetics calculations
To minimize the complications of admixture, we restricted this analysis to the European-American sample. Our model thus has three parameters that we needed to search over: the mutation rate μ, the estimated population size at the start of ARIC, N, and the growth rate during the exponential phase r. We did a grid search over these parameters using the following values: μ: 10−8 per site per generation, up to 9×10−8, in steps of 10−9; r: 1.01 per generation, up to 1.15 per generation in steps of 0.0025; and N: 2×105, up to 4×106, in steps of 105. Because we were comparing with the growth-rate estimate in Gutenkunst et al.,10 of 1.004 per generation, we also separately computed the following likelihoods for r in the vicinity of that, and found them to be negligible.
We fit these parameters to the SFS from our genetic data. To avoid confounding by selection pressures, for this calculation we used an SFS restricted to either sites that were at least 30 base pairs from an exon or third-position sites in codons that could mutate to any nucleotide with no impact on the protein product. We also ignored all sites with genotype calls for fewer than 10,000 people. For sites called in more than 10,000 people, we computed their contributions to the SFS from random samples of 10,000 individuals. We were thus able to assume for this calculation that we had full sample data in exactly 10,000 people at all sites considered.
Our calculation is similar to those described in Wakeley and Takahashi11 and Boyko et al.,12 and is described in the Supplementary Methods.
Additional information
How to cite this article: Coventry, A. et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat. Commun. 1:131 doi: 10.1038/ncomms1130 (2010).
References
- Cohen, J. C. et al. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354, 1264–1272 (2006).
Article CAS PubMed Google Scholar - Fawcett, K. A. et al. Detailed investigation of the role of common and low-frequency WFS1 variants in type 2 diabetes risk. Diabetes 59, 741–746 (2010).
Article CAS PubMed Google Scholar - Glatt, C. et al. Screening a large reference sample to identify very low frequency sequence variants: comparisons between two genes. Nat. Genet. 27, 435–438 (2001).
Article CAS PubMed Google Scholar - Johansen, C. T. et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 42, 684–687 (2010).
Article CAS PubMed PubMed Central Google Scholar - 1000 Genomes. A deep catalog of human genetic variation, http://www.1000genomes.org (2010).
- Personal Genome Project. http://personalgenomes.org (2010).
- Zhang, J. et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput. Biol. 1, e53 (2005).
Article ADS PubMed PubMed Central Google Scholar - Nickerson, D. A. et al. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25, 2745–2751 (1997).
Article CAS PubMed PubMed Central Google Scholar - Stephens, M. et al. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat. Genet. 38, 375–381 (2006).
Article CAS PubMed Google Scholar - Gutenkunst, R. N. et al. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 (2009).
Article PubMed PubMed Central Google Scholar - Wakeley, J. & Takahashi, S. Gene genealogies when the sample size exceeds the effective size of the population. Mol. Biol. Evol. 20, 208–213 (2003).
Article CAS PubMed Google Scholar - Boyko, A. et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4, e1000083 (2008).
Article PubMed PubMed Central Google Scholar - Ramensky, V. et al. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 (2002).
Article CAS PubMed PubMed Central Google Scholar - Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Article CAS PubMed PubMed Central Google Scholar - Arnold, K. et al. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2005).
Article PubMed Google Scholar - Ohta, T. Very slightly deleterious mutations and the molecular clock. J. Mol. Evol. 26, 1–6 (1987).
Article ADS CAS PubMed Google Scholar - Barker, G. Prehistoric Farming in Europe Chapter 10 (Cambridge University Press, 1985).
- Livi-Bacci, M. The Population of Europe (Blackwell, 2000).
- Livi-Bacci, M. A Concise History of World Population (Blackwell, 2001).
- Nachman, M. et al. Estimate of the mutation rate per nucleotide in humans. Genetics 156, 297–304 (2000).
CAS PubMed PubMed Central Google Scholar - Lynch, M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl Acad. Sci. USA 107, 961–968 (2010).
Article ADS CAS PubMed Google Scholar - Hartl, D. & Clark, A. G. Principles of Population Genetics (Sinauer Associates, 2007).
- Levy, S. et al. The diploid genome sequence of an individual human. PLoS Biol. 5, e254 (2007).
Article PubMed PubMed Central Google Scholar - Ng, P. C. et al. Genetic variation in an individual human exome. PLoS Genet. 4, e1000160 (2008).
Article PubMed PubMed Central Google Scholar - Wheeler, D. A. et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876 (2008).
Article ADS CAS PubMed Google Scholar - Wang, J. et al. The diploid genome sequence of an Asian individual. Nature 456, 60–65 (2008).
Article ADS CAS PubMed PubMed Central Google Scholar - Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Article ADS CAS PubMed PubMed Central Google Scholar - Pritchard, J. K. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 69, 124–137 (2001).
Article CAS PubMed PubMed Central Google Scholar - Eyre-Walker, A. Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc. Natl Acad. Sci. USA 107 (suppl 1), 1752–1756 (2010).
Article ADS CAS PubMed Google Scholar - Ewing, B. et al. Base-calling of automated sequencer traces using Phred. Genome Res. 8, 175–194 (1998).
Article CAS PubMed Google Scholar - Altschuler, D. M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Article ADS Google Scholar - Sjölander, K. et al. Dirichlet mixtures: a method for improved detection of weak but significant protein sequence homology. Bioinformatics 12, 327–345 (1996).
Article Google Scholar - Neal, R. Bayesian Learning For Neural Networks (Springer, 1994).
Acknowledgements
We thank Jinghui Zhang for providing source code for SNPDetector7; Phil Green and Brent Ewing for giving permission to distribute the phred30 patch and committing to future distribution of the phred version to which it applies; Kirk Lohmueller for population genetics guidance; Cornelia Scheitz for insights into KCNJ11 and HHEX functional biology; Aida Andres, Nancy Chen, Clement Chow, Tim Connallon, Alon Keinan, Richard Meisel and Margarida Cardoso Moreira for helpful comments on the manuscript; Greg Dyson for useful discussions; and Lawrence S Shimmin, Clement Chow and Peter Schweitzer for insights regarding Sanger trace reading. This work was supported by NIH grant GM065509.
Author information
Author notes
- Alex Coventry and Lara M. Bull-Otterson: These authors contributed equally to this work.
Authors and Affiliations
- Department of Molecular Biology and Genetics, Cornell University, Ithaca, 14853, New York, USA
Alex Coventry & Andrew G. Clark - Department of Molecular & Human Genetics, Human Genome Sequencing Center, Baylor College of Medicine, Houston, 77030, Texas, USA
Lara M. Bull-Otterson, Donna M. Muzny, Lora R. Lewis, David A. Wheeler, Aniko Sabo, Humeira Akbar, Andrew Cree, Alicia C. Hawes, Irene Newsham, Robin T. Varghese, Donna Villasana, Shannon Gross, Vandita Joshi, Jireh Santibanez, Margaret Morgan, Kyle Chang, Walker Hale IV & Richard Gibbs - Human Genetics Center, UT Houston Health Science Center, Houston, 77030, Texas, USA
Xiaoming Liu, Taylor J. Maxwell, Jacy Crosby, James E. Hixson & Eric Boerwinkle - Department of Human Genetics, University of Michigan School of Medicine, Ann Arbor, 48109, Michigan, USA
Thomas J. Rea, Christine Lusk, Kenneth G. Weiss & Charles F. Sing - Department of Biology, Washington University, St Louis, 63130, Missouri, USA
Alan R. Templeton
Authors
- Alex Coventry
You can also search for this author inPubMed Google Scholar - Lara M. Bull-Otterson
You can also search for this author inPubMed Google Scholar - Xiaoming Liu
You can also search for this author inPubMed Google Scholar - Andrew G. Clark
You can also search for this author inPubMed Google Scholar - Taylor J. Maxwell
You can also search for this author inPubMed Google Scholar - Jacy Crosby
You can also search for this author inPubMed Google Scholar - James E. Hixson
You can also search for this author inPubMed Google Scholar - Thomas J. Rea
You can also search for this author inPubMed Google Scholar - Donna M. Muzny
You can also search for this author inPubMed Google Scholar - Lora R. Lewis
You can also search for this author inPubMed Google Scholar - David A. Wheeler
You can also search for this author inPubMed Google Scholar - Aniko Sabo
You can also search for this author inPubMed Google Scholar - Christine Lusk
You can also search for this author inPubMed Google Scholar - Kenneth G. Weiss
You can also search for this author inPubMed Google Scholar - Humeira Akbar
You can also search for this author inPubMed Google Scholar - Andrew Cree
You can also search for this author inPubMed Google Scholar - Alicia C. Hawes
You can also search for this author inPubMed Google Scholar - Irene Newsham
You can also search for this author inPubMed Google Scholar - Robin T. Varghese
You can also search for this author inPubMed Google Scholar - Donna Villasana
You can also search for this author inPubMed Google Scholar - Shannon Gross
You can also search for this author inPubMed Google Scholar - Vandita Joshi
You can also search for this author inPubMed Google Scholar - Jireh Santibanez
You can also search for this author inPubMed Google Scholar - Margaret Morgan
You can also search for this author inPubMed Google Scholar - Kyle Chang
You can also search for this author inPubMed Google Scholar - Walker Hale IV
You can also search for this author inPubMed Google Scholar - Alan R. Templeton
You can also search for this author inPubMed Google Scholar - Eric Boerwinkle
You can also search for this author inPubMed Google Scholar - Richard Gibbs
You can also search for this author inPubMed Google Scholar - Charles F. Sing
You can also search for this author inPubMed Google Scholar
Contributions
The project was conceived by EB, RG and CFS. ARIC samples were obtained by EB. Genes were selected and their variations annotated by TJR, CL, KGW and CFS. Sample preparation and DNA sequencing was done by LMB-O, DMM, LRL, IN, HA, RTV, MM, VJ, JS, SJ, and Andrew Cree. Primary data checking was done by LMB-O and Alex Coventry. The probabilistic SNP calling method was developed by Alex Coventry, who also obtained the primary SNP calls using this algorithm and did the popgen and mutation-rate estimates. The 454 validation was done by LMB-O and AS, with the assistance of HGSC team members DMM, LRL, DAW, HA, Andrew Cree, ACH, IN, RTV, DV, SG, VJ, JS, MM and KC. WH assisted with the release engineering of the genotype caller. The paper was initially drafted by Alex Coventry, LMB-O, AGC and EB, and XL, TJM, TJR, ART, RG and CFS contributed to editing and revising the manuscript. WH assisted in the preparation of the figures.
Corresponding author
Correspondence toAlex Coventry.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Coventry, A., Bull-Otterson, L., Liu, X. et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth.Nat Commun 1, 131 (2010). https://doi.org/10.1038/ncomms1130
- Received: 30 March 2010
- Accepted: 03 November 2010
- Published: 30 November 2010
- DOI: https://doi.org/10.1038/ncomms1130