Improved imputation of common and uncommon SNPs with a new reference set (original) (raw)
- Correspondence
- Published: 27 December 2011
- Kevin B Jacobs1,2,
- Meredith Yeager1,2,
- Amy Hutchinson1,2,
- Joshua Sampson2,
- Nilanjan Chatterjee2,
- Demetrius Albanes2,
- Sonja I Berndt2,
- Charles C Chung2,
- W Ryan Diver3,
- Susan M Gapstur3,
- Lauren R Teras3,
- Christopher A Haiman4,
- Brian E Henderson4,
- Daniel Stram4,
- Xiang Deng1,2,
- Ann W Hsing2,
- Jarmo Virtamo5,
- Michael A Eberle6,
- Jennifer L Stone6,
- Mark P Purdue2,
- Phil Taylor2,
- Margaret Tucker2 &
- …
- Stephen J Chanock2
Nature Genetics volume 44, pages 6–7 (2012)Cite this article
- 914 Accesses
- 39 Citations
- 1 Altmetric
- Metrics details
Statistical imputation of genotype data is an important statistical technique that uses patterns of linkage disequilibrium observed in a reference set of haplotypes to computationally predict genetic variants in silico1. Currently, the most popular reference sets are the publicly available International HapMap2 and 1000 Genomes data sets3. Although these resources are valuable for imputing a sizeable fraction of common SNPs, they may not be optimal for imputing data for the next generation of genome-wide association studies (GWAS) and SNP arrays, which explore a fraction of uncommon variants.
We have built a new resource for the imputation of SNPs for existing and future GWAS, known as the Division of Cancer Epidemiology and Genetics (DCEG) Reference Set. The data set has genotypes for cancer-free individuals, including 728 of European ancestry from three large prospectively sampled studies4,5,6, 98 African-American individuals from the Prostate, Lung, Colon and Ovary Cancer Screening Trial (PLCO), 74 Chinese individuals from a clinical trial in Shanxi, China (SHNX)7 and 349 individuals from the HapMap Project (Table 1). The final harmonized data set includes 2.8 million autosomal polymorphic SNPs for 1,249 individuals after rigorous quality control metrics were applied (see Supplementary Methods and Supplementary Tables 1 and 2).
Table 1 Samples included in the DCEG Reference Set
This is a preview of subscription content, access via your institution
Relevant articles
Open Access articles citing this article.
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
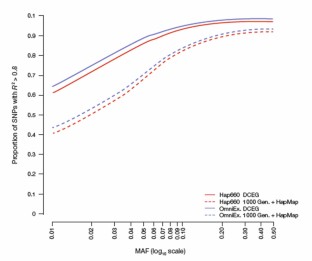
Figure 1: Imputation accuracy for individuals of European ancestry with the DCEG Reference Set and publicly available reference sets.
References
- Marchini, J. & Howie, B. Nat. Rev. Genet. 11, 499–511 (2010).
Article CAS PubMed Google Scholar - Frazer, K.A. et al. Nature 449, 851–861 (2007).
Article CAS PubMed Google Scholar - 1000 Genomes Project Consortium. Nature 467, 1061–1073 (2010).
- Prorok, P.C. et al. Control. Clin. Trials 21, 273S–309S (2000).
Article CAS PubMed Google Scholar - The ATBC Cancer Prevention Study Group. Ann. Epidemiol. 4, 1–10 (1994).
- Calle, E.E. et al. Cancer 94, 2490–2501 (2002).
Article PubMed Google Scholar - Ke, L. Int. J. Cancer 102, 271–274 (2002).
Article CAS PubMed Google Scholar - Howie, B.N., Donnelly, P. & Marchini, J. PLoS Genet. 5, e1000529 (2009).
Article PubMed PubMed Central Google Scholar - Browning, B.L. & Browning, S.R. Am. J. Hum. Genet. 84, 210–223 (2009).
Article CAS PubMed PubMed Central Google Scholar - Park, J.H. et al. Nat. Genet. 42, 570–575 (2010).
Article CAS PubMed PubMed Central Google Scholar - Kolonel, L.N. et al. Am. J. Epidemiol. 151, 346–357 (2000).
Article CAS PubMed Google Scholar
Acknowledgements
The genotyping in the Multiethnic Cohort Study (MEC) was supported by a Department of Defense Breast Cancer Research Program Era of Hope Scholar Award (W81XWH-08-1-0383 to C.A.H.) and an NIH grant (CA132839).
Author information
Authors and Affiliations
- Core Genotyping Facility, SAIC-Frederick, National Cancer Institute (NCI)-Frederick, Frederick, Maryland, USA
Zhaoming Wang, Kevin B Jacobs, Meredith Yeager, Amy Hutchinson & Xiang Deng - Division of Cancer Epidemiology and Genetics, NCI, US National Institutes of Health (NIH), Bethesda, Maryland, USA
Zhaoming Wang, Kevin B Jacobs, Meredith Yeager, Amy Hutchinson, Joshua Sampson, Nilanjan Chatterjee, Demetrius Albanes, Sonja I Berndt, Charles C Chung, Xiang Deng, Ann W Hsing, Mark P Purdue, Phil Taylor, Margaret Tucker & Stephen J Chanock - Epidemiology Research Program, American Cancer Society, Atlanta, Georgia, USA
W Ryan Diver, Susan M Gapstur & Lauren R Teras - Department of Preventive Medicine, Keck School of Medicine, University of Southern California Norris Comprehensive Cancer Center, Los Angeles, California, USA
Christopher A Haiman, Brian E Henderson & Daniel Stram - Department of Chronic Disease Prevention, National Institute for Health and Welfare, Helsinki, Finland
Jarmo Virtamo - Illumina, San Diego, California, USA
Michael A Eberle & Jennifer L Stone
Authors
- Zhaoming Wang
- Kevin B Jacobs
- Meredith Yeager
- Amy Hutchinson
- Joshua Sampson
- Nilanjan Chatterjee
- Demetrius Albanes
- Sonja I Berndt
- Charles C Chung
- W Ryan Diver
- Susan M Gapstur
- Lauren R Teras
- Christopher A Haiman
- Brian E Henderson
- Daniel Stram
- Xiang Deng
- Ann W Hsing
- Jarmo Virtamo
- Michael A Eberle
- Jennifer L Stone
- Mark P Purdue
- Phil Taylor
- Margaret Tucker
- Stephen J Chanock
Corresponding author
Correspondence toStephen J Chanock.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Z., Jacobs, K., Yeager, M. et al. Improved imputation of common and uncommon SNPs with a new reference set.Nat Genet 44, 6–7 (2012). https://doi.org/10.1038/ng.1044
- Published: 27 December 2011
- Issue date: January 2012
- DOI: https://doi.org/10.1038/ng.1044